Somatic TARDBP variants as a cause of semantic dementia
- PMID: 33155043
- PMCID: PMC7805802
- DOI: 10.1093/brain/awaa317
Somatic TARDBP variants as a cause of semantic dementia
Abstract
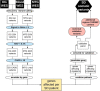
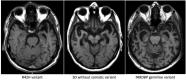
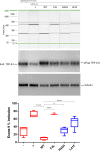
The aetiology of late-onset neurodegenerative diseases is largely unknown. Here we investigated whether de novo somatic variants for semantic dementia can be detected, thereby arguing for a more general role of somatic variants in neurodegenerative disease. Semantic dementia is characterized by a non-familial occurrence, early onset (<65 years), focal temporal atrophy and TDP-43 pathology. To test whether somatic variants in neural progenitor cells during brain development might lead to semantic dementia, we compared deep exome sequencing data of DNA derived from brain and blood of 16 semantic dementia cases. Somatic variants observed in brain tissue and absent in blood were validated using amplicon sequencing and digital PCR. We identified two variants in exon one of the TARDBP gene (L41F and R42H) at low level (1-3%) in cortical regions and in dentate gyrus in two semantic dementia brains, respectively. The pathogenicity of both variants is supported by demonstrating impaired splicing regulation of TDP-43 and by altered subcellular localization of the mutant TDP-43 protein. These findings indicate that somatic variants may cause semantic dementia as a non-hereditary neurodegenerative disease, which might be exemplary for other late-onset neurodegenerative disorders.
Keywords: TARDBP; TDP-43; semantic dementia; somatic variants.
© The Author(s) (2020). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures



References
-
- Barmada SJ, Finkbeiner S.. Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: disease-associated pathways. Rev Neurosci 2010; 21: 251–72. - PubMed
-
- Beck JA, Poulter M, Campbell TA, Uphill JB, Adamson G, Geddes JF, et al.Somatic and germline mosaicism in sporadic early-onset Alzheimer's disease. Hum Mol Genet 2004; 13: 1219–24. - PubMed
-
- Borroni B, Archetti S, Del Bo R, Papetti A, Buratti E, Bonvicini C, et al.TARDBP mutations in frontotemporal lobar degeneration: frequency, clinical features, and disease course. Rejuvenation Res 2010; 13: 509–17. - PubMed
-
- Brady DR, Mufson EJ.. Parvalbumin-immunoreactive neurons in the hippocampal formation of Alzheimer's diseased brain. Neuroscience 1997; 80: 1113–25. - PubMed
-
- Buratti E, Baralle FE.. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 2001; 276: 36337–43. - PubMed

