Drug sensitivity prediction with normal inverse Gaussian shrinkage informed by external data
- PMID: 33155717
- PMCID: PMC7891636
- DOI: 10.1002/bimj.201900371
Drug sensitivity prediction with normal inverse Gaussian shrinkage informed by external data
Abstract
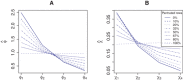
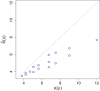
In precision medicine, a common problem is drug sensitivity prediction from cancer tissue cell lines. These types of problems entail modelling multivariate drug responses on high-dimensional molecular feature sets in typically >1000 cell lines. The dimensions of the problem require specialised models and estimation methods. In addition, external information on both the drugs and the features is often available. We propose to model the drug responses through a linear regression with shrinkage enforced through a normal inverse Gaussian prior. We let the prior depend on the external information, and estimate the model and external information dependence in an empirical-variational Bayes framework. We demonstrate the usefulness of this model in both a simulated setting and in the publicly available Genomics of Drug Sensitivity in Cancer data.
Keywords: Genomics of Drug Sensitivity in Cancer (GDSC); drug sensitivity; empirical Bayes; variational Bayes.
© 2020 The Authors. Biometrical Journal published by WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures
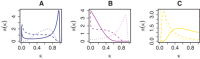
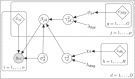
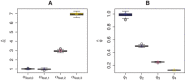


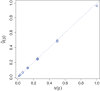
References
-
- Aben, N. , Vis, D. J. , Michaut, M. , & Wessels, L. F. (2016). TANDEM: A two‐stage approach to maximize interpretability of drug response models based on multiple molecular data types. Bioinformatics, 32(17), i413–i420. - PubMed
-
- Ammad‐ud din, M. , Khan, S. A. , Malani, D. , Murumägi, A. , Kallioniemi, O. , Aittokallio, T. , & Kaski, S. (2016). Drug response prediction by inferring pathway‐response associations with kernelized Bayesian matrix factorization. Bioinformatics, 32(17), i455–i463. - PubMed
-
- Barndorff‐Nielsen, O. (1978). Hyperbolic distributions and distributions on hyperbolae. Scandinavian Journal of Statistics, 5(3), 151–157.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

