Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential mycobacterial genes
- PMID: 33155979
- PMCID: PMC7647400
- DOI: 10.7554/eLife.60083
Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential mycobacterial genes
Abstract
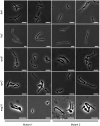
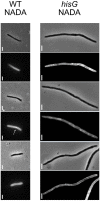
Mycobacterium tuberculosis possesses a large number of genes of unknown or predicted function, undermining fundamental understanding of pathogenicity and drug susceptibility. To address this challenge, we developed a high-throughput functional genomics approach combining inducible CRISPR-interference and image-based analyses of morphological features and sub-cellular chromosomal localizations in the related non-pathogen, M. smegmatis. Applying automated imaging and analysis to 263 essential gene knockdown mutants in an arrayed library, we derive robust, quantitative descriptions of bacillary morphologies consequent on gene silencing. Leveraging statistical-learning, we demonstrate that functionally related genes cluster by morphotypic similarity and that this information can be used to inform investigations of gene function. Exploiting this observation, we infer the existence of a mycobacterial restriction-modification system, and identify filamentation as a defining mycobacterial response to histidine starvation. Our results support the application of large-scale image-based analyses for mycobacterial functional genomics, simultaneously establishing the utility of this approach for drug mechanism-of-action studies.
Keywords: CRISPRi; Mycobacterium smegmatis; Mycobacterium tuberculosis; UMAP; functional genomics; genetics; genomics; infectious disease; microbiology; phenoprint.
Plain language summary
Caused by the microorganism Mycobacterium tuberculosis, tuberculosis kills more people around the world than any other infectious disease. M. tuberculosis is also becoming increasingly resistant to treatments, which are particularly difficult for patients to complete. The M. tuberculosis genome carries about four thousand genes, with several hundred being vital for survival. Finding new ways to fight tuberculosis relies on understanding the exact role of these essential genes, but they are difficult to study in living bacteria. To investigate this question, de Wet et al. used the related, fast-dividing bacterial species called M. smegmatis as a model. Microscopic imaging was combined with CRISPR-interference – a method that temporarily disrupts expression of a specific gene – to examine how blocking an essential gene would affect the shape of the living microorganism. Experiments were conducted on a collection of 270 mutants, capturing single-cell data for hundreds of thousands of live bacteria. To analyze the data, a computational pipeline was built, which automatically clustered similar-shaped bacteria. These groups, or ‘phenoprints’, brought together genes of known and unknown roles; this indicated that these genes participate in similar biological networks – and, if unknown, hinted at their function. Finally, targeting essential genes with CRISPR-interference often yielded the same shape changes as blocking their encoded proteins with antibiotics. This suggests that phenoprints could be useful to understand the mode of action of potential new tuberculosis treatments. When applied to M. tuberculosis and other deadly bacteria, the approach developed by de Wet et al. might speed up drug development.
© 2020, de Wet et al.
Conflict of interest statement
Td, KW, MM, VM, DW No competing interests declared
Figures











Similar articles
-
A Mycobacterial Systems Resource for the Research Community.mBio. 2021 Mar 2;12(2):e02401-20. doi: 10.1128/mBio.02401-20. mBio. 2021. PMID: 33653882 Free PMC article.
-
Dxr is essential in Mycobacterium tuberculosis and fosmidomycin resistance is due to a lack of uptake.BMC Microbiol. 2008 May 20;8:78. doi: 10.1186/1471-2180-8-78. BMC Microbiol. 2008. PMID: 18489786 Free PMC article.
-
[Frontier of mycobacterium research--host vs. mycobacterium].Kekkaku. 2005 Sep;80(9):613-29. Kekkaku. 2005. PMID: 16245793 Japanese.
-
[Development of antituberculous drugs: current status and future prospects].Kekkaku. 2006 Dec;81(12):753-74. Kekkaku. 2006. PMID: 17240921 Review. Japanese.
-
Mycobacterium smegmatis: The Vanguard of Mycobacterial Research.J Bacteriol. 2023 Jan 26;205(1):e0033722. doi: 10.1128/jb.00337-22. Epub 2023 Jan 4. J Bacteriol. 2023. PMID: 36598232 Free PMC article. Review.
Cited by
-
Fluorescence-based CRISPR interference system for controlled genetic repression and live single-cell imaging in mycobacteria.FEBS Lett. 2025 Feb;599(4):488-501. doi: 10.1002/1873-3468.15071. Epub 2024 Dec 1. FEBS Lett. 2025. PMID: 39618159 Free PMC article.
-
The CRISPR-dCas9 interference system suppresses inhA gene expression in Mycobacterium smegmatis.Sci Rep. 2024 Oct 30;14(1):26116. doi: 10.1038/s41598-024-77442-2. Sci Rep. 2024. PMID: 39478003 Free PMC article.
-
Chemical approaches to unraveling the biology of mycobacteria.Cell Chem Biol. 2023 May 18;30(5):420-435. doi: 10.1016/j.chembiol.2023.04.014. Cell Chem Biol. 2023. PMID: 37207631 Free PMC article. Review.
-
Cyclohexyl-griselimycin Is Active against Mycobacterium abscessus in Mice.Antimicrob Agents Chemother. 2022 Jan 18;66(1):e0140021. doi: 10.1128/AAC.01400-21. Epub 2021 Nov 1. Antimicrob Agents Chemother. 2022. PMID: 34723632 Free PMC article.
-
Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.Nat Microbiol. 2023 Oct;8(10):1896-1910. doi: 10.1038/s41564-023-01473-0. Epub 2023 Sep 7. Nat Microbiol. 2023. PMID: 37679597 Free PMC article.
References
-
- Baranowski C, Welsh MA, Sham LT, Eskandarian HA, Lim HC, Kieser KJ, Wagner JC, McKinney JD, Fantner GE, Ioerger TR, Walker S, Bernhardt TG, Rubin EJ, Rego EH. Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife. 2018;7:e37516. doi: 10.7554/eLife.37516. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

