The InterPro protein families and domains database: 20 years on
- PMID: 33156333
- PMCID: PMC7778928
- DOI: 10.1093/nar/gkaa977
The InterPro protein families and domains database: 20 years on
Abstract
The InterPro database (https://www.ebi.ac.uk/interpro/) provides an integrative classification of protein sequences into families, and identifies functionally important domains and conserved sites. InterProScan is the underlying software that allows protein and nucleic acid sequences to be searched against InterPro's signatures. Signatures are predictive models which describe protein families, domains or sites, and are provided by multiple databases. InterPro combines signatures representing equivalent families, domains or sites, and provides additional information such as descriptions, literature references and Gene Ontology (GO) terms, to produce a comprehensive resource for protein classification. Founded in 1999, InterPro has become one of the most widely used resources for protein family annotation. Here, we report the status of InterPro (version 81.0) in its 20th year of operation, and its associated software, including updates to database content, the release of a new website and REST API, and performance improvements in InterProScan.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
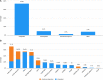


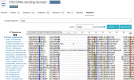
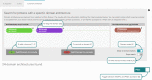
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/S020381/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/T010541/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N019172/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 108433/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- BB/N00521X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

