Cognate base-pair selectivity of hydrophobic unnatural bases in DNA ligation by T4 DNA ligase
- PMID: 33156531
- PMCID: PMC7900958
- DOI: 10.1002/bip.23407
Cognate base-pair selectivity of hydrophobic unnatural bases in DNA ligation by T4 DNA ligase
Abstract
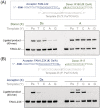
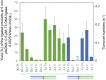
We present cognate base pair selectivity in template-dependent ligation by T4 DNA ligase using a hydrophobic unnatural base pair (UBP), Ds-Pa. T4 DNA ligase efficiently recognizes the Ds-Pa pairing at the conjugation position, and Ds excludes the noncognate pairings with the natural bases. Our results indicate that the hydrophobic base pairing is allowed in enzymatic ligation with higher cognate base-pair selectivity, relative to the hydrogen-bond interactions between pairing bases. The efficient ligation using Ds-Pa can be employed in recombinant DNA technology using genetic alphabet expansion, toward the creation of semi-synthetic organisms containing UBPs.
Keywords: T4 DNA ligase; genetic alphabet expansion; ligation; unnatural base pair.
© 2020 The Authors. Biopolymers published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Karalkar N. B., Benner S. A., Curr. Opin. Chem. Biol. 2018, 46, 188. - PubMed
-
- Lee K. H., Hamashima K., Kimoto M., Hirao I., Curr. Opin. Biotechnol. 2018, 51, 8. - PubMed
-
- Yamashige R., Kimoto M., Okumura R., Hirao I., J. Am. Chem. Soc. 2018, 140, 14038. - PubMed
-
- Moser M. J., Marshall D. J., Grenier J. K., Kieffer C. D., Killeen A. A., Ptacin J. L., Richmond C. S., Roesch E. B., Scherrer C. W., Sherrill C. B., Van Hout C. V., Zanton S. J., Prudent J. R., Clin. Chem. 2003, 49, 407. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

