Auxin-Regulated Reversible Inhibition of TMK1 Signaling by MAKR2 Modulates the Dynamics of Root Gravitropism
- PMID: 33157019
- PMCID: PMC7809621
- DOI: 10.1016/j.cub.2020.10.011
Auxin-Regulated Reversible Inhibition of TMK1 Signaling by MAKR2 Modulates the Dynamics of Root Gravitropism
Abstract
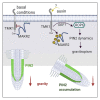
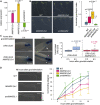
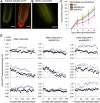
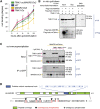
Plants are able to orient their growth according to gravity, which ultimately controls both shoot and root architecture.1 Gravitropism is a dynamic process whereby gravistimulation induces the asymmetric distribution of the plant hormone auxin, leading to asymmetric growth, organ bending, and subsequent reset of auxin distribution back to the original pre-gravistimulation situation.1-3 Differential auxin accumulation during the gravitropic response depends on the activity of polarly localized PIN-FORMED (PIN) auxin-efflux carriers.1-4 In particular, the timing of this dynamic response is regulated by PIN2,5,6 but the underlying molecular mechanisms are poorly understood. Here, we show that MEMBRANE ASSOCIATED KINASE REGULATOR2 (MAKR2) controls the pace of the root gravitropic response. We found that MAKR2 is required for the PIN2 asymmetry during gravitropism by acting as a negative regulator of the cell-surface signaling mediated by the receptor-like kinase TRANSMEMBRANE KINASE1 (TMK1).2,7-10 Furthermore, we show that the MAKR2 inhibitory effect on TMK1 signaling is antagonized by auxin itself, which triggers rapid MAKR2 membrane dissociation in a TMK1-dependent manner. Our findings suggest that the timing of the root gravitropic response is orchestrated by the reversible inhibition of the TMK1 signaling pathway at the cell surface.
Keywords: MAKR; ROP6; TMK; anionic lipids; auxin; brassinosteroid; gravitropism; receptor-like kinase; root.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

References
-
- Su S.H., Gibbs N.M., Jancewicz A.L., Masson P.H. Molecular mechanisms of root gravitropism. Curr. Biol. 2017;27:R964–R972. - PubMed
-
- Rakusová H., Fendrych M., Friml J. Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Curr. Opin. Plant Biol. 2015;23:116–123. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

