The Proteomic Landscape of Centromeric Chromatin Reveals an Essential Role for the Ctf19CCAN Complex in Meiotic Kinetochore Assembly
- PMID: 33157029
- PMCID: PMC7846277
- DOI: 10.1016/j.cub.2020.10.025
The Proteomic Landscape of Centromeric Chromatin Reveals an Essential Role for the Ctf19CCAN Complex in Meiotic Kinetochore Assembly
Abstract
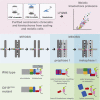
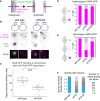
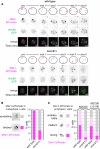
Kinetochores direct chromosome segregation in mitosis and meiosis. Faithful gamete formation through meiosis requires that kinetochores take on new functions that impact homolog pairing, recombination, and the orientation of kinetochore attachment to microtubules in meiosis I. Using an unbiased proteomics pipeline, we determined the composition of centromeric chromatin and kinetochores at distinct cell-cycle stages, revealing extensive reorganization of kinetochores during meiosis. The data uncover a network of meiotic chromosome axis and recombination proteins that bind to centromeres in the absence of the microtubule-binding outer kinetochore sub-complexes during meiotic prophase. We show that the Ctf19cCCAN inner kinetochore complex is essential for kinetochore organization in meiosis. Our functional analyses identify a Ctf19cCCAN-dependent kinetochore assembly pathway that is dispensable for mitotic growth but becomes critical upon meiotic entry. Therefore, changes in kinetochore composition and a distinct assembly pathway specialize meiotic kinetochores for successful gametogenesis.
Keywords: CCAN; Ctf19; budding yeast; centromere; kinetochore; meiosis; metaphase I; prophase I; proteomics.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





Similar articles
-
Functional characterization of kinetochore protein, Ctf19 in meiosis I: an implication of differential impact of Ctf19 on the assembly of mitotic and meiotic kinetochores in Saccharomyces cerevisiae.Mol Microbiol. 2014 Mar;91(6):1179-99. doi: 10.1111/mmi.12527. Epub 2014 Feb 19. Mol Microbiol. 2014. PMID: 24446862
-
The kinetochore prevents centromere-proximal crossover recombination during meiosis.Elife. 2015 Dec 14;4:e10850. doi: 10.7554/eLife.10850. Elife. 2015. PMID: 26653857 Free PMC article.
-
The structure of the Ctf19c/CCAN from budding yeast.Elife. 2019 Feb 14;8:e44239. doi: 10.7554/eLife.44239. Elife. 2019. PMID: 30762520 Free PMC article.
-
Critical Foundation of the Kinetochore: The Constitutive Centromere-Associated Network (CCAN).Prog Mol Subcell Biol. 2017;56:29-57. doi: 10.1007/978-3-319-58592-5_2. Prog Mol Subcell Biol. 2017. PMID: 28840232 Review.
-
Kinetochore composition and its function: lessons from yeasts.FEMS Microbiol Rev. 2014 Mar;38(2):185-200. doi: 10.1111/1574-6976.12049. FEMS Microbiol Rev. 2014. PMID: 24666101 Review.
Cited by
-
Centromeres: From chromosome biology to biotechnology applications and synthetic genomes in plants.Plant Biotechnol J. 2022 Nov;20(11):2051-2063. doi: 10.1111/pbi.13875. Epub 2022 Jul 7. Plant Biotechnol J. 2022. PMID: 35722725 Free PMC article. Review.
-
Centromere pairing enables correct segregation of meiotic chromosomes.Curr Biol. 2024 May 20;34(10):2085-2093.e6. doi: 10.1016/j.cub.2024.04.008. Epub 2024 Apr 25. Curr Biol. 2024. PMID: 38670094 Free PMC article.
-
Use of Time-Lapse Microscopy and Stage-Specific Nuclear Depletion of Proteins to Study Meiosis in S. Cerevisiae.J Vis Exp. 2022 Oct 11;(188):10.3791/64580. doi: 10.3791/64580. J Vis Exp. 2022. PMID: 36314815 Free PMC article.
-
Biochemical characterisation of Mer3 helicase interactions and the protection of meiotic recombination intermediates.Nucleic Acids Res. 2023 May 22;51(9):4363-4384. doi: 10.1093/nar/gkad175. Nucleic Acids Res. 2023. PMID: 36942481 Free PMC article.
-
The Spo13/Meikin pathway confines the onset of gamete differentiation to meiosis II in yeast.EMBO J. 2022 Feb 15;41(4):e109446. doi: 10.15252/embj.2021109446. Epub 2022 Jan 13. EMBO J. 2022. PMID: 35023198 Free PMC article.
References
-
- Hinshaw S.M., Harrison S.C. Kinetochore function from the bottom up. Trends Cell Biol. 2018;28:22–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

