Genes causing congenital hydrocephalus: Their chromosomal characteristics of telomere proximity and DNA compositions
- PMID: 33157092
- PMCID: PMC7750280
- DOI: 10.1016/j.expneurol.2020.113523
Genes causing congenital hydrocephalus: Their chromosomal characteristics of telomere proximity and DNA compositions
Abstract
Congenital hydrocephalus (CH) is caused by genetic mutations, but whether factors impacting human genetic mutations are disease-specific remains elusive. Given two factors associated with high mutation rates, we reviewed how many disease-susceptible genes match with (i) proximity to telomeres or (ii) high adenine and thymine (A + T) content in human CH as compared to other disorders of the central nervous system (CNS). We extracted genomic information using a genome data viewer. Importantly, 98 of 108 genes causing CH satisfied (i) or (ii), resulting in >90% matching rate. However, such a high accordance no longer sustained as we checked two factors in Alzheimer's disease (AD) and/or familial Parkinson's disease (fPD), resulting in 84% and 59% matching, respectively. A disease-specific matching of telomere proximity or high A + T content predicts causative genes of CH much better than neurodegenerative diseases and other CNS conditions, likely due to sufficient number of known causative genes (n = 108) and precise determination and classification of the genotype and phenotype. Our analysis suggests a need for identifying genetic basis of both factors before human clinical studies, to prioritize putative genes found in preclinical models into the likely (meeting at least one) and more likely candidate (meeting both), which predisposes human genes to mutations.
Keywords: Alzheimer's disease; A + T content, mutation; Chromosome, homologous recombination, familial Parkinson's disease; Congenital hydrocephalus; Telomeres.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest
The authors declare no competing financial interests.
Figures


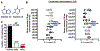


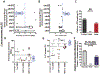
References
-
- Alazami AM, Maddirevula S, Seidahmed MZ, Albhlal LA, Alkuraya FS, 2019. A novel ISLR2-linked autosomal recessive syndrome of congenital hydrocephalus, arthrogryposis and abdominal distension. Hum. Genet 138, 105–107. - PubMed
-
- Albright AL, Ferson S, Carlos S, 2005. Occult hydrocephalus in children with cerebral palsy. Neurosurgery 56, 93–96 (discussion 96-97). - PubMed
-
- Alby C, Piquand K, Huber C, Megarbane A, Ichkou A, Legendre M, Pelluard F, Encha-Ravazi F, Abi-Tayeh G, Bessieres B, El Chehadeh-Djebbar S, Laurent N, Faivre L, Sztriha L, Zombor M, Szabo H, Failler M, Garfa-Traore M, Bole C, Nitschke P, Nizon M, Elkhartoufi N, Clerget-Darpoux F, Munnich A, Lyonnet S, Vekemans M, Saunier S, Cormier-Daire V, Attie-Bitach T, Thomas S, 2015. Mutations in KIAA0586 Cause Lethal Ciliopathies Ranging from a Hydrolethalus Phenotype to Short-Rib Polydactyly Syndrome. Am J Hum Genet 97, 311–318. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

