Portable Rabies Virus Sequencing in Canine Rabies Endemic Countries Using the Oxford Nanopore MinION
- PMID: 33158200
- PMCID: PMC7694271
- DOI: 10.3390/v12111255
Portable Rabies Virus Sequencing in Canine Rabies Endemic Countries Using the Oxford Nanopore MinION
Abstract
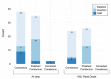
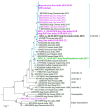
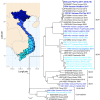
As countries with endemic canine rabies progress towards elimination by 2030, it will become necessary to employ techniques to help plan, monitor, and confirm canine rabies elimination. Sequencing can provide critical information to inform control and vaccination strategies by identifying genetically distinct virus variants that may have different host reservoir species or geographic distributions. However, many rabies testing laboratories lack the resources or expertise for sequencing, especially in remote or rural areas where human rabies deaths are highest. We developed a low-cost, high throughput rabies virus sequencing method using the Oxford Nanopore MinION portable sequencer. A total of 259 sequences were generated from diverse rabies virus isolates in public health laboratories lacking rabies virus sequencing capacity in Guatemala, India, Kenya, and Vietnam. Phylogenetic analysis provided valuable insight into rabies virus diversity and distribution in these countries and identified a new rabies virus lineage in Kenya, the first published canine rabies virus sequence from Guatemala, evidence of rabies spread across an international border in Vietnam, and importation of a rabid dog into a state working to become rabies-free in India. Taken together, our evaluation highlights the MinION's potential for low-cost, high volume sequencing of pathogens in locations with limited resources.
Keywords: MinION; canine rabies elimination; lyssavirus; nanopore; portable sequencing; rabies.
Conflict of interest statement
The sponsors had no role in the design, execution, interpretation, or writing of the study.
Figures


References
-
- Virus Taxonomy: 2019 Release. [(accessed on 19 October 2020)]; Available online: https://talk.ictvonline.org/taxonomy/
-
- World Health Organization . WHO Expert Consultation on Rabies: Third Report. World Health Organization; Geneva, Switzerland: 2018.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

