Optical Biosensors for the Detection of Rheumatoid Arthritis (RA) Biomarkers: A Comprehensive Review
- PMID: 33158306
- PMCID: PMC7663853
- DOI: 10.3390/s20216289
Optical Biosensors for the Detection of Rheumatoid Arthritis (RA) Biomarkers: A Comprehensive Review
Abstract
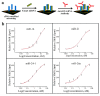
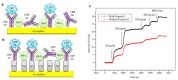
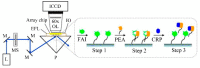
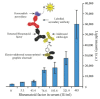
A comprehensive review of optical biosensors for the detection of biomarkers associated with rheumatoid arthritis (RA) is presented here, including microRNAs (miRNAs), C-reactive protein (CRP), rheumatoid factor (RF), anti-citrullinated protein antibodies (ACPA), interleukin-6 (IL-6) and histidine, which are biomarkers that enable RA detection and/or monitoring. An overview of the different optical biosensors (based on fluorescence, plasmon resonances, interferometry, surface-enhanced Raman spectroscopy (SERS) among other optical techniques) used to detect these biomarkers is given, describing their performance and main characteristics (limit of detection (LOD) and dynamic range), as well as the connection between the respective biomarker and rheumatoid arthritis. It has been observed that the relationship between the corresponding biomarker and rheumatoid arthritis tends to be obviated most of the time when explaining the mechanism of the optical biosensor, which forces the researcher to look for further information about the biomarker. This review work attempts to establish a clear association between optical sensors and rheumatoid arthritis biomarkers as well as to be an easy-to-use tool for the researchers working in this field.
Keywords: CRP; biomarkers; miRNA; optical biosensors; rheumatoid arthritis (RA).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
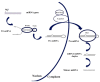

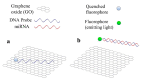
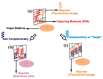
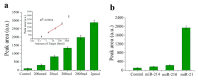




References
-
- Duarte-Garcia A., reviewed by American Collegue of Rheumatology Diseases & Conditions. Rheumatoid Arthritis. [(accessed on 20 October 2020)]; Available online: https://www.rheumatology.org/I-Am-A/Patient-Caregiver/Diseases-Condition....
-
- Driver C.B., Stöppler M.C. Rheumatoid Arthritis (RA): Early Signs, Symptoms, Causes, Treatment & Diet. [(accessed on 5 May 2020)]; Available online: https://www.emedicinehealth.com/rheumatoid_arthritis/article_em.htm#rheu....
-
- England B.R., Tiong B.K., Bergman M.J., Curtis J.R., Kazi S., Mikuls T.R., O’Dell J.R., Ranganath V.K., Limanni A., Suter L.G., et al. 2019 Update of the American College of Rheumatology Recommended Rheumatoid Arthritis Disease Activity Measures. Arthritis Care Res. 2019;71:1540–1555. doi: 10.1002/acr.24042. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

