Genetically Defined, Syngeneic Organoid Platform for Developing Combination Therapies for Ovarian Cancer
- PMID: 33158842
- PMCID: PMC7858239
- DOI: 10.1158/2159-8290.CD-20-0455
Genetically Defined, Syngeneic Organoid Platform for Developing Combination Therapies for Ovarian Cancer
Abstract
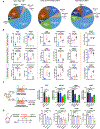
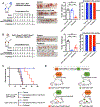
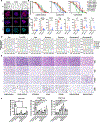
The paucity of genetically informed, immunocompetent tumor models impedes evaluation of conventional, targeted, and immune therapies. By engineering mouse fallopian tube epithelial organoids using lentiviral gene transduction and/or CRISPR/Cas9 mutagenesis, we generated multiple high-grade serous tubo-ovarian cancer (HGSC) models exhibiting mutational combinations seen in patients with HGSC. Detailed analysis of homologous recombination (HR)-proficient (Trp53-/-;Ccne1OE;Akt2OE;KrasOE ), HR-deficient (Trp53-/-;Brca1-/-;MycOE ), and unclassified (Trp53-/-;Pten-/-;Nf1-/- ) organoids revealed differences in in vitro properties (proliferation, differentiation, and "secretome"), copy-number aberrations, and tumorigenicity. Tumorigenic organoids had variable sensitivity to HGSC chemotherapeutics, and evoked distinct immune microenvironments that could be modulated by neutralizing organoid-produced chemokines/cytokines. These findings enabled development of a chemotherapy/immunotherapy regimen that yielded durable, T cell-dependent responses in Trp53-/-;Ccne1OE;Akt2OE;Kras HGSC; in contrast, Trp53-/-;Pten-/-;Nf1-/- tumors failed to respond. Mouse and human HGSC models showed genotype-dependent similarities in chemosensitivity, secretome, and immune microenvironment. Genotype-informed, syngeneic organoid models could provide a platform for the rapid evaluation of tumor biology and therapeutics. SIGNIFICANCE: The lack of genetically informed, diverse, immunocompetent models poses a major barrier to therapeutic development for many malignancies. Using engineered fallopian tube organoids to study the cell-autonomous and cell-nonautonomous effects of specific combinations of mutations found in HGSC, we suggest an effective combination treatment for the currently intractable CCNE1-amplified subgroup.This article is highlighted in the In This Issue feature, p. 211.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

