The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase
- PMID: 33158944
- PMCID: PMC7925111
- DOI: 10.1128/JVI.01969-20
The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase
Abstract
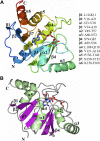
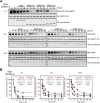
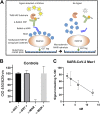
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and other SARS-related CoVs encode 3 tandem macrodomains within nonstructural protein 3 (nsp3). The first macrodomain, Mac1, is conserved throughout CoVs and binds to and hydrolyzes mono-ADP-ribose (MAR) from target proteins. Mac1 likely counters host-mediated antiviral ADP-ribosylation, a posttranslational modification that is part of the host response to viral infections. Mac1 is essential for pathogenesis in multiple animal models of CoV infection, implicating it as a virulence factor and potential therapeutic target. Here, we report the crystal structure of SARS-CoV-2 Mac1 in complex with ADP-ribose. SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) Mac1 domains exhibit similar structural folds, and all 3 proteins bound to ADP-ribose with affinities in the low micromolar range. Importantly, using ADP-ribose-detecting binding reagents in both a gel-based assay and novel enzyme-linked immunosorbent assays (ELISAs), we demonstrated de-MARylating activity for all 3 CoV Mac1 proteins, with the SARS-CoV-2 Mac1 protein leading to a more rapid loss of substrate than the others. In addition, none of these enzymes could hydrolyze poly-ADP-ribose. We conclude that the SARS-CoV-2 and other CoV Mac1 proteins are MAR-hydrolases with similar functions, indicating that compounds targeting CoV Mac1 proteins may have broad anti-CoV activity.IMPORTANCE SARS-CoV-2 has recently emerged into the human population and has led to a worldwide pandemic of COVID-19 that has caused more than 1.2 million deaths worldwide. With no currently approved treatments, novel therapeutic strategies are desperately needed. All coronaviruses encode a highly conserved macrodomain (Mac1) that binds to and removes ADP-ribose adducts from proteins in a dynamic posttranslational process that is increasingly being recognized as an important factor that regulates viral infection. The macrodomain is essential for CoV pathogenesis and may be a novel therapeutic target. Thus, understanding its biochemistry and enzyme activity are critical first steps for these efforts. Here, we report the crystal structure of SARS-CoV-2 Mac1 in complex with ADP-ribose and describe its ADP-ribose binding and hydrolysis activities in direct comparison to those of SARS-CoV and MERS-CoV Mac1 proteins. These results are an important first step for the design and testing of potential therapies targeting this unique protein domain.
Keywords: ADP-ribose; SARS-CoV-2; coronavirus; macrodomain; mono-ADP-ribose; poly-ADP-ribose.
Copyright © 2021 Alhammad et al.
Figures








Update of
-
The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase.bioRxiv [Preprint]. 2020 Oct 28:2020.05.11.089375. doi: 10.1101/2020.05.11.089375. bioRxiv. 2020. Update in: J Virol. 2021 Jan 13;95(3):e01969-20. doi: 10.1128/JVI.01969-20. PMID: 32511412 Free PMC article. Updated. Preprint.
References
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

