A single-cell atlas and lineage analysis of the adult Drosophila ovary
- PMID: 33159074
- PMCID: PMC7648648
- DOI: 10.1038/s41467-020-19361-0
A single-cell atlas and lineage analysis of the adult Drosophila ovary
Erratum in
-
Author Correction: A single-cell atlas and lineage analysis of the adult Drosophila ovary.Nat Commun. 2021 Oct 6;12(1):5951. doi: 10.1038/s41467-021-26191-1. Nat Commun. 2021. PMID: 34615857 Free PMC article. No abstract available.
Abstract
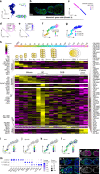
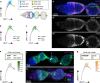
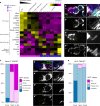
The Drosophila ovary is a widely used model for germ cell and somatic tissue biology. Here we use single-cell RNA-sequencing (scRNA-seq) to build a comprehensive cell atlas of the adult Drosophila ovary that contains transcriptional profiles for every major cell type in the ovary, including the germline stem cells and their niche cells, follicle stem cells, and previously undescribed subpopulations of escort cells. In addition, we identify Gal4 lines with specific expression patterns and perform lineage tracing of subpopulations of escort cells and follicle cells. We discover that a distinct subpopulation of escort cells is able to convert to follicle stem cells in response to starvation or upon genetic manipulation, including knockdown of escargot, or overactivation of mTor or Toll signalling.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Xie T, Spradling A. C. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. - PubMed
-
- Margolis J, Spradling A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 1995;121:3797–3807. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

