SARS-CoV-2: Comparative analysis of different RNA extraction methods
- PMID: 33160015
- PMCID: PMC7640895
- DOI: 10.1016/j.jviromet.2020.114008
SARS-CoV-2: Comparative analysis of different RNA extraction methods
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent of the COVID-19 pandemic. Although other diagnostic methods have been introduced, detection of viral genes on oro- and nasopharyngeal swabs by reverse-transcription real time-PCR (rRT-PCR) assays is still the gold standard. Efficient viral RNA extraction is a prerequisite for downstream performance of rRT-PCR assays. Currently, several automatic methods that include RNA extraction are available. However, due to the growing demand, a shortage in kit supplies could be experienced in several labs. For these reasons, the use of different commercial or in-house protocols for RNA extraction may increase the possibility to analyze high number of samples. Herein, we compared the efficiency of RNA extraction of three different commercial kits and an in-house extraction protocol using synthetic ssRNA standards of SARS-CoV-2 as well as in oro-nasopharyngeal swabs from six COVID-19-positive patients. It was concluded that tested commercial kits can be used with some modifications for the detection of the SARS-CoV-2 genome by rRT-PCR approaches, although with some differences in RNA yields. Conversely, EXTRAzol reagent was the less efficient due to the phase separation principle at the basis of RNA extraction. Overall, this study offers alternative suitable methods to manually extract RNA that can be taken into account for SARS-CoV-2 detection.
Keywords: Oro- nasopharyngeal swabs; RNA extraction; SARS-CoV-2; rRT-PCR.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
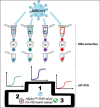
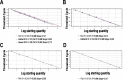
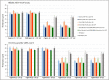
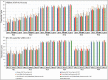
References
-
- CDC: Center for Disease Control and Prevention. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

