Distinct Stage Changes in Early-Life Colonization and Acquisition of the Gut Microbiota and Its Correlations With Volatile Fatty Acids in Goat Kids
- PMID: 33162961
- PMCID: PMC7581860
- DOI: 10.3389/fmicb.2020.584742
Distinct Stage Changes in Early-Life Colonization and Acquisition of the Gut Microbiota and Its Correlations With Volatile Fatty Acids in Goat Kids
Abstract
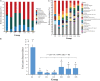
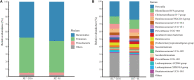
In livestock, a comprehensive understanding of the early-life establishment and acquisition of commensal gut microbiota allow us to develop better husbandry management operations and manipulate the gut microbiota for young animals, improving the efficiency of animal production. Here, we collected 123 microbial samples of 11 healthy goat kids and their mothers to investigate the colonization and acquisition of the gut microbiota and their correlations with volatile fatty acids (VFAs) in goat kids from birth to day 56. An age-dependent increasing and more homogeneous diversity were observed for the feces of goat kids. Overall, Firmicutes, Bacteroidetes, and Proteobacteria were the predominant phyla in the fecal microbiota of goat kids, but their relative abundance varied considerably with age. Accordingly, the colonization of the fecal microbiota in goat kids was divided into three distinct stages: newborn (day 0), non-rumination stage (days 7-21), and transition stages (days 28-56). LEfSe analysis revealed a total of 49 bacterial biomarkers that are stage-specific (LDA score > 3, P < 0.05). Significant Spearman correlations (P < 0.05) were observed between the abundances of several bacterial biomarkers and the VFA concentrations. Furthermore, a substantial difference in the fecal microbiota composition was present between 56-day-old goat kids and mothers, whereas there was a moderate difference in the rumen microbiota between them. Among four body sites (i.e., feces, oral cavity, vagina, and breast milk) of mothers, the maternal vaginal and breast milk microbiota were the major source of the fecal microbiota of goat kids in the first 56 days after birth, although their contributions decreased with age and unknown sources increased after day 28. In summary, we concluded that the gut bacterial community in goat kids after birth was mainly acquired from the maternal vagina and breast milk. Its colonization showed three distinct phases with dramatic shifts of composition mainly driven by age and diet changes. Our results provide a framework for a better understanding of the roles of the gut microbiota in young ruminants.
Keywords: acquisition; early life; establishment; fecal microbiota; goat; rumen microbiota; volatile fatty acid.
Copyright © 2020 Guo, Li, Zhang, Zhang, Wang, Li and Zhang.
Figures



References
-
- Abecia L., Ramosmorales E., Martinezfernandez G., Arco A., Martingarcia A. I., Newbold C. J., et al. (2014). Feeding management in early life influences microbial colonisation and fermentation in the rumen of newborn goat kids. Anim. Prod. Sci. 54 1449–1454.
LinkOut - more resources
Full Text Sources

