A Novel In-Cell ELISA Assay Allows Rapid and Automated Quantification of SARS-CoV-2 to Analyze Neutralizing Antibodies and Antiviral Compounds
- PMID: 33162987
- PMCID: PMC7581787
- DOI: 10.3389/fimmu.2020.573526
A Novel In-Cell ELISA Assay Allows Rapid and Automated Quantification of SARS-CoV-2 to Analyze Neutralizing Antibodies and Antiviral Compounds
Abstract
The coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently the most pressing medical and socioeconomic challenge. Constituting important correlates of protection, the determination of virus-neutralizing antibodies (NAbs) is indispensable for convalescent plasma selection, vaccine candidate evaluation, and immunity certificates. In contrast to standard serological ELISAs, plaque reduction neutralization tests (PRNTs) are laborious, time-consuming, expensive, and restricted to specialized laboratories. To replace microscopic counting-based SARS-CoV-2 PRNTs by a novel assay exempt from genetically modified viruses, which are inapplicable in most diagnostics departments, we established a simple, rapid, and automated SARS-CoV-2 neutralization assay employing an in-cell ELISA (icELISA) approach. After optimization of various parameters such as virus-specific antibodies, cell lines, virus doses, and duration of infection, SARS-CoV-2-infected cells became amenable as direct antigen source for quantitative icELISA. Antiviral agents such as human sera containing NAbs or antiviral interferons dose dependently reduced the SARS-CoV-2-specific signal. Applying increased infectious doses, the icELISA-based neutralization test (icNT) was superior to PRNT in discriminating convalescent sera with high from those with intermediate neutralizing capacities. In addition, the icNT was found to be specific, discriminating between SARS-CoV-2-specific NAbs and those raised against other coronaviruses. Altogether, the SARS-CoV-2 icELISA test allows rapid (<48 h in total, read-out in seconds) and automated quantification of virus infection in cell culture to evaluate the efficacy of NAbs and antiviral drugs using reagents and equipment present in most routine diagnostics departments.
Keywords: COVID-19; SARS-CoV-2; antibodies; interferon; neutralizing; vaccine.
Copyright © 2020 Schöler, Le-Trilling, Eilbrecht, Mennerich, Anastasiou, Krawczyk, Herrmann, Dittmer and Trilling.
Figures

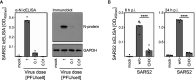
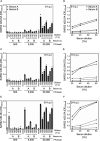


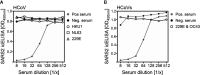
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

