Efficient Sequencing, Assembly, and Annotation of Human KIR Haplotypes
- PMID: 33162997
- PMCID: PMC7581912
- DOI: 10.3389/fimmu.2020.582927
Efficient Sequencing, Assembly, and Annotation of Human KIR Haplotypes
Abstract
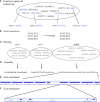
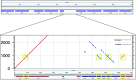
The homology, recombination, variation, and repetitive elements in the natural killer-cell immunoglobulin-like receptor (KIR) region has made full haplotype DNA interpretation impossible in a high-throughput workflow. Here, we present a new approach using long-read sequencing to efficiently capture, sequence, and assemble diploid human KIR haplotypes. Probes were designed to capture KIR fragments efficiently by leveraging the repeating homology of the region. IDT xGen® Lockdown probes were used to capture 2-8 kb of sheared DNA fragments followed by sequencing on a PacBio Sequel. The sequences were error corrected, binned, and then assembled using the Canu assembler. The location of genes and their exon/intron boundaries are included in the workflow. The assembly and annotation was evaluated on 16 individuals (8 African American and 8 Europeans) from whom ground truth was known via long-range sequencing with fosmid library preparation. Using only 18 capture probes, the results show that the assemblies cover 97% of the GenBank reference, are 99.97% concordant, and it takes only 1.8 haplotigs to cover 75% of the reference. We also report the first assembly of diploid KIR haplotypes from long-read WGS. Our targeted hybridization probe capture and sequencing approach is the first of its kind to fully sequence and phase all diploid human KIR haplotypes, and it is efficient enough for population-scale studies and clinical use. The open and free software is available at https://github.com/droeatumn/kass and supported by a environment at https://hub.docker.com/repository/docker/droeatumn/kass.
Keywords: DNA; annotation; assembly; haplotype; killer-cell immunoglobulin-like receptor; natural killer.
Copyright © 2020 Roe, Williams, Ivery, Brouckaert, Downey, Locklear, Kuang and Maiers.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

