Genome-wide analysis of PHD finger gene family and identification of potential miRNA and their PHD finger gene specific targets in Oryza sativa indica
- PMID: 33163736
- PMCID: PMC7610035
- DOI: 10.1016/j.ncrna.2020.10.002
Genome-wide analysis of PHD finger gene family and identification of potential miRNA and their PHD finger gene specific targets in Oryza sativa indica
Abstract
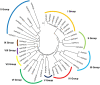
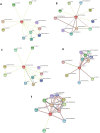
Rice (Oryza sativa L.) is one of the most important cereal crops for one third of the world population. However, the grain quality as well as yield of rice is severely affected by various abiotic stresses. Environmental stresses affect the expression of various microRNAs (miRNAs) which in turn negatively regulate gene expression at the post-transcriptional level either by degrading the target mRNA genes or suppressing translation in plants. Plant homeo-domain (PHD) finger proteins are known to be involved in the plant response to salinity stress. In the present study, we identified 44 putative OsPHD finger genes in Oryza sativa Indica, using Ensembl Plants Database. Using computational approach, potential miRNAs that target OsPHD finger genes were identified. Out of the 44 OsPHD finger genes only three OsPHD finger genes i.e., OsPHD2, OsPHD35 and OsPHD11, were found to be targeted by five newly identified putative miRNAs i.e., ath-miRf10010-akr, ath-miRf10110-akr, osa-miR1857-3p, osa-miRf10863-akr, and osa-miRf11806-akr. This is the first report of these five identified miRNAs on targeting PHD finger in Oryza sativa Indica. Further, expression analysis of 44 PHD finger genes under salinity was also performed using quantitative Real-Time PCR. The expression profile of 8 genes were found to be differentially regulated, among them two genes were significantly up regulated i.e., OsPHD6 and OsPHD12. In silico protein-protein interaction analysis using STRING database showed interaction of the OsPHD finger proteins with other protein partners that are directly or indirectly involved in development and abiotic stress tolerance.
Keywords: Gene expression; Oryza sativa; PHD finger; Protein-protein interaction; Salinity stress; Transcription factor; microRNA.
© 2020 [The Author/The Authors].
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





References
-
- Mohammadi-Nejad G., Singh R.K., Arzani A., Rezaie A.M., Sabouri H., Gregorio G.B. Evaluation of salinity tolerance in rice genotypes. Int. J. Plant Prod. 2010;4(3):199–208.
-
- Sabouri H., Sabouri A. New evidence of QTLs attributed to salinity tolerance in rice. Afr. J. Biotechnol. 2008;7(24):4376–4383.
-
- Islam M.R., Salam M.A., Hassan L., Collard B.C.Y., Singh R.K., Gregorio G.B. QTL mapping for salinity tolerance at seedling stage in rice. Emir. J. Food Agric. 2011;23(2):137–146.
-
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
LinkOut - more resources
Full Text Sources

