A prognostic model for overall survival of patients with early-stage non-small cell lung cancer: a multicentre, retrospective study
- PMID: 33163952
- PMCID: PMC7646741
- DOI: 10.1016/s2589-7500(20)30225-9
A prognostic model for overall survival of patients with early-stage non-small cell lung cancer: a multicentre, retrospective study
Abstract
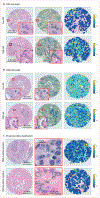
Background: Intratumoural heterogeneity has been previously shown to be related to clonal evolution and genetic instability and associated with tumour progression. Phenotypically, it is reflected in the diversity of appearance and morphology within cell populations. Computer-extracted features relating to tumour cellular diversity on routine tissue images might correlate with outcome. This study investigated the prognostic ability of computer-extracted features of tumour cellular diversity (CellDiv) from haematoxylin and eosin (H&E)-stained histology images of non-small cell lung carcinomas (NSCLCs).
Methods: In this multicentre, retrospective study, we included 1057 patients with early-stage NSCLC with corresponding diagnostic histology slides and overall survival information from four different centres. CellDiv features quantifying local cellular morphological diversity from H&E-stained histology images were extracted from the tumour epithelium region. A Cox proportional hazards model based on CellDiv was used to construct risk scores for lung adenocarcinoma (LUAD; 270 patients) and lung squamous cell carcinoma (LUSC; 216 patients) separately using data from two of the cohorts, and was validated in the two remaining independent cohorts (comprising 236 patients with LUAD and 335 patients with LUSC). We used multivariable Cox regression analysis to examine the predictive ability of CellDiv features for 5-year overall survival, controlling for the effects of clinical and pathological parameters. We did a gene set enrichment and Gene Ontology analysis on 405 patients to identify associations with differentially expressed biological pathways implicated in lung cancer pathogenesis.
Findings: For prognosis of patients with early-stage LUSC, the CellDiv LUSC model included 11 discriminative CellDiv features, whereas for patients with early-stage LUAD, the model included 23 features. In the independent validation cohorts, patients predicted to be at a higher risk by the univariable CellDiv model had significantly worse 5-year overall survival (hazard ratio 1·48 [95% CI 1·06-2·08]; p=0·022 for The Cancer Genome Atlas [TCGA] LUSC group, 2·24 [1·04-4·80]; p=0·039 for the University of Bern LUSC group, and 1·62 [1·15-2·30]; p=0·0058 for the TCGA LUAD group). The identified CellDiv features were also found to be strongly associated with apoptotic signalling and cell differentiation pathways.
Interpretation: CellDiv features were strongly prognostic of 5-year overall survival in patients with early-stage NSCLC and also associated with apoptotic signalling and cell differentiation pathways. The CellDiv-based risk stratification model could potentially help to determine which patients with early-stage NSCLC might receive added benefit from adjuvant therapy.
Funding: National Institue of Health and US Department of Defense.
Conflict of interest statement
Declaration of interests CL, KB, and AM have a pending patent (Predicting Cancer Recurrence Using Local Co-occurrence of Cell Morphology). AM is an equity holder in Elucid Bioimaging and in Inspirata, to whom his technology has been licensed. He is currently a scientific advisory board member at Aiforia. He is also involved in a National Institutes of Health U24 grant with PathCore and is involved in four grants with Inspirata. His work has received sponsored research funding from Bristol Myers Squibb, AstraZeneca, and Philips, outside of the submitted work. AJ has patents 10528848 and 9111179 issued, and patents 20190266726, 20190251687, 20180129911, and 20160307305 pending (US patents, registered in Case Western Reserve University). VV reports grants and personal fees from Merck, Bristol Myers Squibb, Genentech, AstraZeneca, Celgene, Novartis, Amgen, Fulgent Genetics, Reddy Labs, Alkermes, Nektar Therapeutics, Novocure, and Foundation Medicine, outside of the submitted work; and advisory or consulting fees from Genentech, Merck, Bristol Myers Squibb, AstraZeneca, Foundation Medicine, Nektar Therapeutics, Alkermes, Reddy Labs, and Millennium Pharma, outside of the submitted work. All other authors declare no competing interests.
Figures



References
-
- Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol Mech Dis 2013; 8: 277–302. - PubMed
-
- Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus 2014; 37: E11. - PubMed
-
- Yuan Y, Failmezger H, Rueda OM, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 2012; 4: 157ra143. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA216579/CA/NCI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- C06 RR012463/RR/NCRR NIH HHS/United States
- IK6 BX006185/BX/BLRD VA/United States
- U24 CA199374/CA/NCI NIH HHS/United States
- R43 EB028736/EB/NIBIB NIH HHS/United States
- U01 CA239055/CA/NCI NIH HHS/United States
- R01 CA249992/CA/NCI NIH HHS/United States
- R01 CA220581/CA/NCI NIH HHS/United States
- P50 CA196530/CA/NCI NIH HHS/United States
- R01 CA202752/CA/NCI NIH HHS/United States
- R01 CA208236/CA/NCI NIH HHS/United States
- U01 CA248226/CA/NCI NIH HHS/United States
- I01 BX004121/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
