IS26-mediated amplification of blaOXA-1 and blaCTX-M-15 with concurrent outer membrane porin disruption associated with de novo carbapenem resistance in a recurrent bacteraemia cohort
- PMID: 33164081
- PMCID: PMC7816169
- DOI: 10.1093/jac/dkaa447
IS26-mediated amplification of blaOXA-1 and blaCTX-M-15 with concurrent outer membrane porin disruption associated with de novo carbapenem resistance in a recurrent bacteraemia cohort
Abstract
Background: Approximately half of clinical carbapenem-resistant Enterobacterales (CRE) isolates lack carbapenem-hydrolysing enzymes and develop carbapenem resistance through alternative mechanisms.
Objectives: To elucidate development of carbapenem resistance mechanisms from clonal, recurrent ESBL-positive Enterobacterales (ESBL-E) bacteraemia isolates in a vulnerable patient population.
Methods: This study investigated a cohort of ESBL-E bacteraemia cases in Houston, TX, USA. Oxford Nanopore Technologies long-read and Illumina short-read sequencing data were used for comparative genomic analysis. Serial passaging experiments were performed on a set of clinical ST131 Escherichia coli isolates to recapitulate in vivo observations. Quantitative PCR (qPCR) and qRT-PCR were used to determine copy number and transcript levels of β-lactamase genes, respectively.
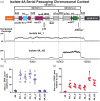
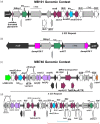
Results: Non-carbapenemase-producing CRE (non-CP-CRE) clinical isolates emerged from an ESBL-E background through a concurrence of primarily IS26-mediated amplifications of blaOXA-1 and blaCTX-M-1 group genes coupled with porin inactivation. The discrete, modular translocatable units (TUs) that carried and amplified β-lactamase genes mobilized intracellularly from a chromosomal, IS26-bound transposon and inserted within porin genes, thereby increasing β-lactamase gene copy number and inactivating porins concurrently. The carbapenem resistance phenotype and TU-mediated β-lactamase gene amplification were recapitulated by passaging a clinical ESBL-E isolate in the presence of ertapenem. Clinical non-CP-CRE isolates had stable carbapenem resistance phenotypes in the absence of ertapenem exposure.
Conclusions: These data demonstrate IS26-mediated mechanisms underlying β-lactamase gene amplification with concurrent outer membrane porin disruption driving emergence of clinical non-CP-CRE. Furthermore, these amplifications were stable in the absence of antimicrobial pressure. Long-read sequencing can be utilized to identify unique mobile genetic element mechanisms that drive antimicrobial resistance.
© The Author(s) 2020. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



References
-
- CDC. Antibiotic Resistance Threats in the United States 2019. 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-re...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

