Integration of genomics and transcriptomics predicts diabetic retinopathy susceptibility genes
- PMID: 33164750
- PMCID: PMC7728435
- DOI: 10.7554/eLife.59980
Integration of genomics and transcriptomics predicts diabetic retinopathy susceptibility genes
Abstract
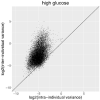
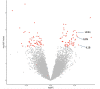
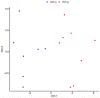
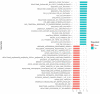
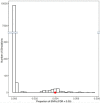
We determined differential gene expression in response to high glucose in lymphoblastoid cell lines derived from matched individuals with type 1 diabetes with and without retinopathy. Those genes exhibiting the largest difference in glucose response were assessed for association with diabetic retinopathy in a genome-wide association study meta-analysis. Expression quantitative trait loci (eQTLs) of the glucose response genes were tested for association with diabetic retinopathy. We detected an enrichment of the eQTLs from the glucose response genes among small association p-values and identified folliculin (FLCN) as a susceptibility gene for diabetic retinopathy. Expression of FLCN in response to glucose was greater in individuals with diabetic retinopathy. Independent cohorts of individuals with diabetes revealed an association of FLCN eQTLs with diabetic retinopathy. Mendelian randomization confirmed a direct positive effect of increased FLCN expression on retinopathy. Integrating genetic association with gene expression implicated FLCN as a disease gene for diabetic retinopathy.
Keywords: LCLs; diabetic retinopathy; eQTL; folliculin; gene expression; genetics; genomics; human; mendelian randomization.
Plain language summary
One of the side effects of diabetes is loss of vision from diabetic retinopathy, which is caused by injury to the light sensing tissue in the eye, the retina. Almost all individuals with diabetes develop diabetic retinopathy to some extent, and it is the leading cause of irreversible vision loss in working-age adults in the United States. How long a person has been living with diabetes, the extent of increased blood sugars and genetics all contribute to the risk and severity of diabetic retinopathy. Unfortunately, virtually no genes associated with diabetic retinopathy have yet been identified. When a gene is activated, it produces messenger molecules known as mRNA that are used by cells as instructions to produce proteins. The analysis of mRNA molecules, as well as genes themselves, can reveal the role of certain genes in disease. The studies of all genes and their associated mRNAs are respectively called genomics and transcriptomics. Genomics reveals what genes are present, while transcriptomics shows how active genes are in different cells. Skol et al. developed methods to study genomics and transcriptomics together to help discover genes that cause diabetic retinopathy. Genes involved in how cells respond to high blood sugar were first identified using cells grown in the lab. By comparing the activity of these genes in people with and without retinopathy the study identified genes associated with an increased risk of retinopathy in diabetes. In people with retinopathy, the activity of the folliculin gene (FLCN) increased more in response to high blood sugar. This was further verified with independent groups of people and using computer models to estimate the effect of different versions of the folliculin gene. The methods used here could be applied to understand complex genetics in other diseases. The results provide new understanding of the effects of diabetes. They may also help in the development of new treatments for diabetic retinopathy, which are likely to improve on the current approach of using laser surgery or injections into the eye.
© 2020, Skol et al.
Conflict of interest statement
AS, SJ, AS, SC, SF, OS, PB, AL, MS, DC, AS, IB, BS, MG No competing interests declared
Figures






References
-
- Arar NH, Freedman BI, Adler SG, Iyengar SK, Chew EY, Davis MD, Satko SG, Bowden DW, Duggirala R, Elston RC, Guo X, Hanson RL, Igo RP, Ipp E, Kimmel PL, Knowler WC, Molineros J, Nelson RG, Pahl MV, Quade SRE, Rasooly RS, Rotter JI, Saad MF, Scavini M, Schelling JR, Sedor JR, Shah VO, Zager PG, Abboud HE. Heritability of the severity of diabetic retinopathy: the FIND-Eye study. Investigative Opthalmology & Visual Science. 2008;49:3839–3845. doi: 10.1167/iovs.07-1633. - DOI - PMC - PubMed
-
- Becker RA. The New S Language. Wadsworth & Brooks/Cole; 1988.
-
- Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, Strathdee CA, Dempster J, Lyons NJ, Burns R, Nag A, Kugener G, Cimini B, Tsvetkov P, Maruvka YE, O'Rourke R, Garrity A, Tubelli AA, Bandopadhayay P, Tsherniak A, Vazquez F, Wong B, Birger C, Ghandi M, Thorner AR, Bittker JA, Meyerson M, Getz G, Beroukhim R, Golub TR. Genetic and transcriptional evolution alters Cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Dryad/10.5061/dryad.zkh18938j
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

