Practitioner's Guide to Latent Class Analysis: Methodological Considerations and Common Pitfalls
- PMID: 33165028
- PMCID: PMC7746621
- DOI: 10.1097/CCM.0000000000004710
Practitioner's Guide to Latent Class Analysis: Methodological Considerations and Common Pitfalls
Abstract
Latent class analysis is a probabilistic modeling algorithm that allows clustering of data and statistical inference. There has been a recent upsurge in the application of latent class analysis in the fields of critical care, respiratory medicine, and beyond. In this review, we present a brief overview of the principles behind latent class analysis. Furthermore, in a stepwise manner, we outline the key processes necessary to perform latent class analysis including some of the challenges and pitfalls faced at each of these steps. The review provides a one-stop shop for investigators seeking to apply latent class analysis to their data.
Copyright © 2020 by the Society of Critical Care Medicine and Wolters Kluwer Health, Inc. All Rights Reserved.
Figures


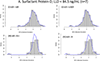



References
-
- Marshall JC: Why have clinical trials in sepsis failed? Trends Mol Med 2014; 20(4):195–203 - PubMed
-
- Soni N: ARDS, acronyms and the Pinocchio effect. Anaesthesia 2010; 65(10):976–979 - PubMed
-
- Pavord ID, Beasley R, Agusti A, Anderson GP, et al. : After asthma: redefining airways diseases. Lancet 2018; 391(10118):350–400 - PubMed

