Network medicine in Cardiovascular Research
- PMID: 33165538
- PMCID: PMC8404464
- DOI: 10.1093/cvr/cvaa321
Network medicine in Cardiovascular Research
Abstract
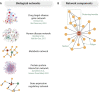
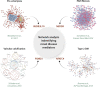
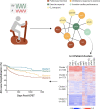
The ability to generate multi-omics data coupled with deeply characterizing the clinical phenotype of individual patients promises to improve understanding of complex cardiovascular pathobiology. There remains an important disconnection between the magnitude and granularity of these data and our ability to improve phenotype-genotype correlations for complex cardiovascular diseases. This shortcoming may be due to limitations associated with traditional reductionist analytical methods, which tend to emphasize a single molecular event in the pathogenesis of diseases more aptly characterized by crosstalk between overlapping molecular pathways. Network medicine is a rapidly growing discipline that considers diseases as the consequences of perturbed interactions between multiple interconnected biological components. This powerful integrative approach has enabled a number of important discoveries in complex disease mechanisms. In this review, we introduce the basic concepts of network medicine and highlight specific examples by which this approach has accelerated cardiovascular research. We also review how network medicine is well-positioned to promote rational drug design for patients with cardiovascular diseases, with particular emphasis on advancing precision medicine.
Keywords: Cardiovascular disease; Network medicine; Omics; Pathobiology; Precision medicine.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author(s) 2020. For permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Diez D, Wheelock AM, Goto S, Haeggstrom JZ, Paulsson-Berne G, Hansson GK, Hedin U, Gabrielsen A, Wheelock CE.. The use of network analyses for elucidating mechanisms in cardiovascular disease. Mol Biosyst 2010;6:289–304. - PubMed

