Analysis of the small chromosomal Prionium serratum (Cyperid) demonstrates the importance of reliable methods to differentiate between mono- and holocentricity
- PMID: 33165742
- PMCID: PMC7665975
- DOI: 10.1007/s00412-020-00745-6
Analysis of the small chromosomal Prionium serratum (Cyperid) demonstrates the importance of reliable methods to differentiate between mono- and holocentricity
Abstract
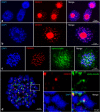
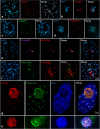
For a long time, the Cyperid clade (Thurniceae-Juncaceae-Cyperaceae) was considered a group of species possessing holocentromeres exclusively. The basal phylogenetic position of Prionium serratum (Thunb.) Drège (Thurniceae) within Cyperids makes this species an important specimen to understand the centromere evolution within this clade. In contrast to the expectation, the chromosomal distribution of the centromere-specific histone H3 (CENH3), alpha-tubulin and different centromere-associated post-translational histone modifications (H3S10ph, H3S28ph and H2AT120ph) demonstrate a monocentromeric organisation of P. serratum chromosomes. Analysis of the high-copy repeat composition resulted in the identification of two centromere-localised satellite repeats. Hence, monocentricity was the ancestral condition for the Juncaceae-Cyperaceae-Thurniaceae Cyperid clade, and holocentricity in this clade has independently arisen at least twice after differentiation of the three families, once in Juncaceae and the other one in Cyperaceae. In this context, methods suitable for the identification of holocentromeres are discussed.
Keywords: CENH3/CENPA; Centromere type; Cyperids; Evolution; Holocentric chromosome; Thurniceae.
Figures



Similar articles
-
Chromosome size matters: genome evolution in the cyperid clade.Ann Bot. 2022 Dec 31;130(7):999-1014. doi: 10.1093/aob/mcac136. Ann Bot. 2022. PMID: 36342743 Free PMC article.
-
The holocentricity in the dioecious nutmeg (Myristica fragrans) is not based on major satellite repeats.Chromosome Res. 2024 May 8;32(2):8. doi: 10.1007/s10577-024-09751-1. Chromosome Res. 2024. PMID: 38717688 Free PMC article.
-
Repeat-based holocentromeres of the woodrush Luzula sylvatica reveal insights into the evolutionary transition to holocentricity.Nat Commun. 2024 Nov 5;15(1):9565. doi: 10.1038/s41467-024-53944-5. Nat Commun. 2024. PMID: 39500889 Free PMC article.
-
Plant centromeres: genetics, epigenetics and evolution.Mol Biol Rep. 2018 Oct;45(5):1491-1497. doi: 10.1007/s11033-018-4284-7. Epub 2018 Aug 16. Mol Biol Rep. 2018. PMID: 30117088 Review.
-
Holocentromere identity: from the typical mitotic linear structure to the great plasticity of meiotic holocentromeres.Chromosoma. 2016 Sep;125(4):669-81. doi: 10.1007/s00412-016-0612-7. Epub 2016 Aug 16. Chromosoma. 2016. PMID: 27530342 Review.
Cited by
-
Chromosomal evolution in Cryptangieae Benth. (Cyperaceae): Evidence of holocentrism and pseudomonads.Protoplasma. 2024 May;261(3):527-541. doi: 10.1007/s00709-023-01915-w. Epub 2023 Dec 21. Protoplasma. 2024. PMID: 38123818
-
Meiotic recombination dynamics in plants with repeat-based holocentromeres shed light on the primary drivers of crossover patterning.Nat Plants. 2024 Mar;10(3):423-438. doi: 10.1038/s41477-024-01625-y. Epub 2024 Feb 9. Nat Plants. 2024. PMID: 38337039 Free PMC article.
-
Chromosome size matters: genome evolution in the cyperid clade.Ann Bot. 2022 Dec 31;130(7):999-1014. doi: 10.1093/aob/mcac136. Ann Bot. 2022. PMID: 36342743 Free PMC article.
-
The holocentricity in the dioecious nutmeg (Myristica fragrans) is not based on major satellite repeats.Chromosome Res. 2024 May 8;32(2):8. doi: 10.1007/s10577-024-09751-1. Chromosome Res. 2024. PMID: 38717688 Free PMC article.
-
Immunodetection of tubulin and centromeric histone H3 (CENH3) proteins in Glycine species.Mol Biol Rep. 2024 Jul 13;51(1):792. doi: 10.1007/s11033-024-09730-z. Mol Biol Rep. 2024. PMID: 39001981
References
-
- Aliyeva-Schnorr L, Beier S, Karafiatova M, Schmutzer T, Scholz U, Dolezel J, Stein N, Houben A. Cytogenetic mapping with centromeric bacterial artificial chromosomes contigs shows that this recombination-poor region comprises more than half of barley chromosome 3H. Plant J. 2015;84:385–394. - PubMed
-
- Andrews S (2010) FastQC: A quality control tool for high throughput sequence data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Bass HW, Hoffman GG, Lee TJ, Wear EE, Joseph SR, Allen GC, Hanley-Bowdoin L, Thompson WF. Defining multiple, distinct, and shared spatiotemporal patterns of DNA replication and endoreduplication from 3D image analysis of developing maize (Zea mays L.) root tip nuclei. Plant Mol Biol. 2015;89:339–351. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

