Optimization and evaluation of propolis liposomes as a promising therapeutic approach for COVID-19
- PMID: 33166584
- PMCID: PMC7647905
- DOI: 10.1016/j.ijpharm.2020.120028
Optimization and evaluation of propolis liposomes as a promising therapeutic approach for COVID-19
Abstract
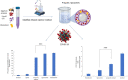
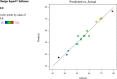
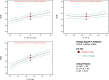
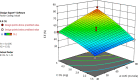
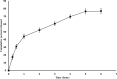
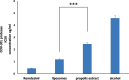
The present work aimed to develop an optimized liposomal formulation for enhancing the anti-viral activity of propolis against COVID-19. Docking studies were performed for certain components of Egyptian Propolis using Avigan, Hydroxychloroquine and Remdesivir as standard antivirals against both COVID-19 3CL-protease and S1 spike protein. Response surface methodology and modified injection method were implemented to maximize the entrapment efficiency and release of the liposomal formulation. The optimized formulation parameters were as follow: LMC of 60 mM, CH% of 20% and DL of 5 mg/ml. At those values the E.E% and released % were 70.112% and 81.801%, respectively with nanosized particles (117 ± 11 nm). Docking studies revealed that Rutin and Caffeic acid phenethyl ester showed the highest affinity to both targets. Results showed a significant inhibitory effect of the optimized liposomal formula of Propolis against COVID-3CL protease (IC50 = 1.183 ± 0.06) compared with the Egyptian propolis extract (IC50 = 2.452 ± 0.11), P < 0.001. Interestingly, the inhibition of viral replication of COVID-19 determined by RT_PCR has been significantly enhanced via encapsulation of propolis extract within the liposomal formulation (P < 0.0001) and was comparable to the viral inhibitory effect of the potent antiviral (remdesivir). These findings identified the potential of propolis liposomes as a promising treatment approach against COVID-19.
Keywords: 3CL-protease; COVID-19; Liposomes; Propolis; RT-PCR; Spike protein.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Ghaffar K., Marasini N., Giddam A., Batzloff M., Good M., Skwarczynski M., Toth I. The Role of Size in Development of Mucosal Liposome-Lipopeptide Vaccine Candidates Against Group A Streptococcus. MC. 2016;13(1):22–27. - PubMed
-
- Abagyan R., Totrov M. Biased Probability Monte Carlo Conformational Searches and Electrostatic Calculations for Peptides and Proteins. Journal of Molecular Biology. 1994;235(3):983–1002. - PubMed
-
- Akaji K., Konno H., Mitsui H., Teruya K., Shimamoto Y., Hattori Y., Ozaki T., Kusunoki M., Sanjoh A. Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors. J. Med. Chem. 2011;54(23):7962–7973. - PubMed
-
- Badria F., Fathy H., Fatehe A., Ahmed M., Ghazy M. Chemical and biological diversity of propolis samples from Bulgaria, Libya and Egypt. J Apither. 2018;4(1):17. doi: 10.5455/ja.10.5455/ja.20180428014109. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

