Confidence Sets for Cohen's d effect size images
- PMID: 33166643
- PMCID: PMC7836238
- DOI: 10.1016/j.neuroimage.2020.117477
Confidence Sets for Cohen's d effect size images
Abstract
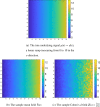
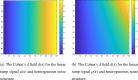
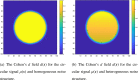
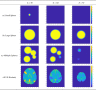
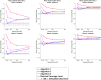
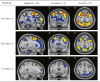
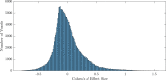
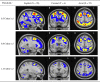
Current statistical inference methods for task-fMRI suffer from two fundamental limitations. First, the focus is solely on detection of non-zero signal or signal change, a problem that is exacerbated for large scale studies (e.g. UK Biobank, N=40,000+) where the 'null hypothesis fallacy' causes even trivial effects to be determined as significant. Second, for any sample size, widely used cluster inference methods only indicate regions where a null hypothesis can be rejected, without providing any notion of spatial uncertainty about the activation. In this work, we address these issues by developing spatial Confidence Sets (CSs) on clusters found in thresholded Cohen's d effect size images. We produce an upper and lower CS to make confidence statements about brain regions where Cohen's d effect sizes have exceeded and fallen short of a non-zero threshold, respectively. The CSs convey information about the magnitude and reliability of effect sizes that is usually given separately in a t-statistic and effect estimate map. We expand the theory developed in our previous work on CSs for %BOLD change effect maps (Bowring et al., 2019) using recent results from the bootstrapping literature. By assessing the empirical coverage with 2D and 3D Monte Carlo simulations resembling fMRI data, we find our method is accurate in sample sizes as low as N=60. We compute Cohen's d CSs for the Human Connectome Project working memory task-fMRI data, illustrating the brain regions with a reliable Cohen's d response for a given threshold. By comparing the CSs with results obtained from a traditional statistical voxelwise inference, we highlight the improvement in activation localization that can be gained with the Confidence Sets.
Keywords: Cohen’s d; Confidence sets; Effect sizes; Task fmri; fMRI.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures














References
-
- Alfaro-Almagro F., Jenkinson M., Bangerter N.K., Andersson J.L.R., Griffanti L., Douaud G., Sotiropoulos S.N., Jbabdi S., Hernandez-Fernandez M., Vallee E., Vidaurre D., Webster M., McCarthy P., Rorden C., Daducci A., Alexander D.C., Zhang H., Dragonu I., Matthews P.M., Miller K.L., Smith S.M. Image processing and quality control for the first 10,000 brain imaging datasets from UK biobank. NeuroImage. 2018;166:400–424. - PMC - PubMed
-
- Button K.S., Ioannidis J.P.A., Mokrysz C., Nosek B.A., Flint J., Robinson E.S.J., Munafò M.R. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14(5):365–376. - PubMed
-
- Carp J. The secret lives of experiments: methods reporting in the fMRI literature. NeuroImage. 2012;63(1):289–300. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

