Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy
- PMID: 33167030
- PMCID: PMC7736788
- DOI: 10.1093/nar/gkaa1013
Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy
Erratum in
-
Correction to 'Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy'.Nucleic Acids Res. 2021 Jul 9;49(12):7204-7205. doi: 10.1093/nar/gkab568. Nucleic Acids Res. 2021. PMID: 34161581 Free PMC article. No abstract available.
Abstract
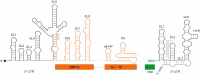
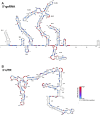
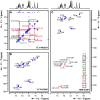
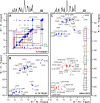
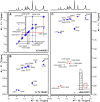
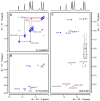
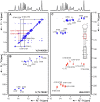
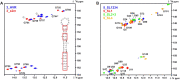
The current pandemic situation caused by the Betacoronavirus SARS-CoV-2 (SCoV2) highlights the need for coordinated research to combat COVID-19. A particularly important aspect is the development of medication. In addition to viral proteins, structured RNA elements represent a potent alternative as drug targets. The search for drugs that target RNA requires their high-resolution structural characterization. Using nuclear magnetic resonance (NMR) spectroscopy, a worldwide consortium of NMR researchers aims to characterize potential RNA drug targets of SCoV2. Here, we report the characterization of 15 conserved RNA elements located at the 5' end, the ribosomal frameshift segment and the 3'-untranslated region (3'-UTR) of the SCoV2 genome, their large-scale production and NMR-based secondary structure determination. The NMR data are corroborated with secondary structure probing by DMS footprinting experiments. The close agreement of NMR secondary structure determination of isolated RNA elements with DMS footprinting and NMR performed on larger RNA regions shows that the secondary structure elements fold independently. The NMR data reported here provide the basis for NMR investigations of RNA function, RNA interactions with viral and host proteins and screening campaigns to identify potential RNA binders for pharmaceutical intervention.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

