Development of Diagnostic Tests for Detection of SARS-CoV-2
- PMID: 33167445
- PMCID: PMC7694548
- DOI: 10.3390/diagnostics10110905
Development of Diagnostic Tests for Detection of SARS-CoV-2
Abstract
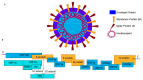
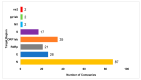
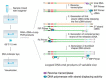
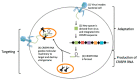
One of the most effective ways to prevent the spread of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is to develop accurate and rapid diagnostic tests. There are a number of molecular, serological, and imaging methods that are used to diagnose this infection in hospitals and clinical settings. The purpose of this review paper is to present the available approaches for detecting SARS-CoV-2 and address the advantages and limitations of each detection method. This work includes studies from recent literature publications along with information from the manufacturer's manuals of commercially available SARS-CoV-2 diagnostic products. Furthermore, supplementary information from the Food & Drug Administration (FDA), Centers for Disease Control and Prevention (CDC), and World Health Organization (WHO) is cited. The viral components targeted for virus detection, the principles of each diagnostic technique, and the detection efficiency of each approach are discussed. The potential of using diagnostic tests that were originally developed for previous epidemic viruses is also presented.
Keywords: COVID-19; SARS-CoV-2; diagnostic tests; disease detection; viral infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

