Histone Variants: Guardians of Genome Integrity
- PMID: 33167489
- PMCID: PMC7694513
- DOI: 10.3390/cells9112424
Histone Variants: Guardians of Genome Integrity
Abstract
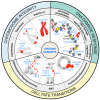
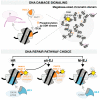
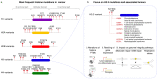
Chromatin integrity is key for cell homeostasis and for preventing pathological development. Alterations in core chromatin components, histone proteins, recently came into the spotlight through the discovery of their driving role in cancer. Building on these findings, in this review, we discuss how histone variants and their associated chaperones safeguard genome stability and protect against tumorigenesis. Accumulating evidence supports the contribution of histone variants and their chaperones to the maintenance of chromosomal integrity and to various steps of the DNA damage response, including damaged chromatin dynamics, DNA damage repair, and damage-dependent transcription regulation. We present our current knowledge on these topics and review recent advances in deciphering how alterations in histone variant sequence, expression, and deposition into chromatin fuel oncogenic transformation by impacting cell proliferation and cell fate transitions. We also highlight open questions and upcoming challenges in this rapidly growing field.
Keywords: DNA damage response; DNA repair; cancer; cell fate; chromatin; chromosome integrity; genome stability; histone chaperones; histone variants; oncohistones.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

