Histone H3K9 Demethylase JMJD2B Plays a Role in LXRα-Dependent Lipogenesis
- PMID: 33167594
- PMCID: PMC7664202
- DOI: 10.3390/ijms21218313
Histone H3K9 Demethylase JMJD2B Plays a Role in LXRα-Dependent Lipogenesis
Abstract
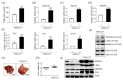
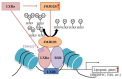
Ligand-activated liver X receptor α (LXRα) upregulates the expression of hepatic lipogenic genes, which leads to triglyceride (TG) accumulation, resulting in nonalcoholic fatty liver disease (NAFLD). Thus, LXRα regulation may provide a novel therapeutic target against NAFLD. However, histone methylation-mediated epigenetic regulation involved in LXRα-dependent lipogenesis is poorly understood. In this study, we investigated the functional role of the histone demethylase Jumonji domain-containing protein 2B (JMJD2B) in LXRα-dependent lipogenesis. JMJD2B expression level was upregulated in HepG2 cells treated with LXRα agonist T0901317 or palmitate and the liver of mice administered with T0901317 or fed a high-fat diet. Knockdown of JMJD2B using siRNA abrogated T0901317-induced LXRα-dependent lipogenic gene expression and lowered intracellular TG accumulation. Conversely, overexpression of JMJD2B in HepG2 cells upregulated the expression of LXRα-dependent lipogenic genes, in line with increased intracellular TG levels. JMJD2B overexpression or T0901317 treatment induced the recruitment of JMJD2B and LXRα to LXR response elements (LXRE) in the promoter region of LXRα-target gene and reduced the enrichment of H3K9me2 and H3K9me3 in the vicinity of the LXRE. Furthermore, JMJD2B enhanced T0901317 or LXRα-induced transcriptional activities of reporters containing LXRE. A co-immunoprecipitation assay revealed that JMJD2B interacted with activated LXRα. Moreover, overexpression of JMJD2B in mice resulted in upregulation of hepatic LXRα-dependent lipogenic genes, consistent with development of hepatic steatosis. Taken together, these results indicate that JMJD2B plays a role in LXRα-mediated lipogenesis via removing the repressive histone marks, H3K9me2 and H3K9me3, at LXRE, which might contribute to hepatic steatosis.
Keywords: hepatic steatosis; histone demethylase Jumonji domain-containing protein 2B; histone methylation; ligand-activated liver X receptor α; lipogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

