A nuclear transport-related gene signature combined with IDH mutation and 1p/19q codeletion better predicts the prognosis of glioma patients
- PMID: 33167941
- PMCID: PMC7654069
- DOI: 10.1186/s12885-020-07552-3
A nuclear transport-related gene signature combined with IDH mutation and 1p/19q codeletion better predicts the prognosis of glioma patients
Abstract
Background: The nuclear transport system has been proposed to be indispensable for cell proliferation and invasion in cancers. Prognostic biomarkers and molecular targets in nuclear transport systems have been developed. However, no systematic analysis of genes related to nuclear transport in gliomas has been performed. An integrated prognostic classification involving mutation and nuclear transport gene signatures has not yet been explored.
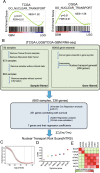
Methods: In the present study, we analyzed gliomas from a training cohort (TCGA dataset, n = 660) and validation cohort (CGGA dataset, n = 668) to develop a prognostic nuclear transport gene signature and generate an integrated classification system. Gene set enrichment analysis (GSEA) showed that glioblastoma (GBM) was mainly enriched in nuclear transport progress compared to lower-grade glioma (LGG). Then, we developed a nuclear transport risk score (NTRS) for gliomas with a training cohort. NTRS was significantly correlated with clinical and genetic characteristics, including grade, age, histology, IDH status and 1p/19q codeletion, in the training and validation cohorts.
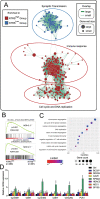
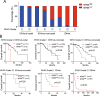
Results: Survival analysis revealed that patients with a higher NTRS exhibited shorter overall survival. NTRS showed better prognostic value compared to classical molecular markers, including IDH status and 1p/19q codeletion. Furthermore, univariate and multivariate analyses indicated that NTRS was an independent prognostic factor for gliomas. Enrichment map and Gene Ontology analysis demonstrated that signaling pathways related to the cell cycle were enriched in the NTRSHigh group. Subgroup survival analysis revealed that NTRS could differentiate the outcomes of low- and high-risk patients with wild-type IDH or mutant IDH and 1p/19q non-codeletion.
Conclusions: NTRS is associated with poor outcomes and could be an independent prognostic marker in diffuse gliomas. Prognostic classification combined with IDH mutation, 1p/19q codeletion and NTRS could better predict the survival of glioma patients.
Keywords: CGGA; Classification; Gene signature; Gliomas; Nuclear transport; Prognosis; TCGA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Garg M, Kanojia D, Mayakonda A, Ganesan TS, Sadhanandhan B, Suresh S, Nagare RP, Said JW, Doan NB, et al. Selinexor (KPT-330) has antitumor activity against anaplastic thyroid carcinoma in vitro and in vivo and enhances sensitivity to doxorubicin. Sci Rep. 2017;7(1):9749. doi: 10.1038/s41598-017-10325-x. - DOI - PMC - PubMed
-
- Rodriguez-Bravo V, Pippa R, Song WM, Carceles-Cordon M, Dominguez-Andres A, Fujiwara N, Woo J, Koh AP, Ertel A, Lokareddy RK, et al. Nuclear pores promote lethal prostate Cancer by increasing POM121-driven E2F1, MYC, and AR nuclear import. Cell. 2018;174(5):1200–1215. doi: 10.1016/j.cell.2018.07.015. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

