Deciphering cell-cell interactions and communication from gene expression
- PMID: 33168968
- PMCID: PMC7649713
- DOI: 10.1038/s41576-020-00292-x
Deciphering cell-cell interactions and communication from gene expression
Abstract
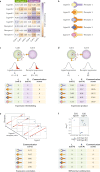
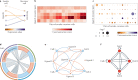
Cell-cell interactions orchestrate organismal development, homeostasis and single-cell functions. When cells do not properly interact or improperly decode molecular messages, disease ensues. Thus, the identification and quantification of intercellular signalling pathways has become a common analysis performed across diverse disciplines. The expansion of protein-protein interaction databases and recent advances in RNA sequencing technologies have enabled routine analyses of intercellular signalling from gene expression measurements of bulk and single-cell data sets. In particular, ligand-receptor pairs can be used to infer intercellular communication from the coordinated expression of their cognate genes. In this Review, we highlight discoveries enabled by analyses of cell-cell interactions from transcriptomic data and review the methods and tools used in this context.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

