c-Rel orchestrates energy-dependent epithelial and macrophage reprogramming in fibrosis
- PMID: 33168981
- PMCID: PMC7116435
- DOI: 10.1038/s42255-020-00306-2
c-Rel orchestrates energy-dependent epithelial and macrophage reprogramming in fibrosis
Erratum in
-
Author Correction: c-Rel orchestrates energy-dependent epithelial and macrophage reprogramming in fibrosis.Nat Metab. 2021 Jan;3(1):118-119. doi: 10.1038/s42255-020-00326-y. Nat Metab. 2021. PMID: 33303984 No abstract available.
Abstract
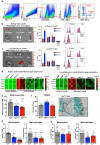
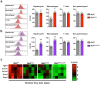
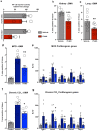
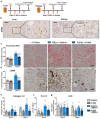
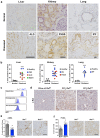
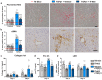
Fibrosis is a common pathological feature of chronic disease. Deletion of the NF-κB subunit c-Rel limits fibrosis in multiple organs, although the mechanistic nature of this protection is unresolved. Using cell-specific gene-targeting manipulations in mice undergoing liver damage, we elucidate a critical role for c-Rel in controlling metabolic changes required for inflammatory and fibrogenic activities of hepatocytes and macrophages and identify Pfkfb3 as the key downstream metabolic mediator of this response. Independent deletions of Rel in hepatocytes or macrophages suppressed liver fibrosis induced by carbon tetrachloride, while combined deletion had an additive anti-fibrogenic effect. In transforming growth factor-β1-induced hepatocytes, c-Rel regulates expression of a pro-fibrogenic secretome comprising inflammatory molecules and connective tissue growth factor, the latter promoting collagen secretion from HMs. Macrophages lacking c-Rel fail to polarize to M1 or M2 states, explaining reduced fibrosis in RelΔLysM mice. Pharmacological inhibition of c-Rel attenuated multi-organ fibrosis in both murine and human fibrosis. In conclusion, activation of c-Rel/Pfkfb3 in damaged tissue instigates a paracrine signalling network among epithelial, myeloid and mesenchymal cells to stimulate fibrogenesis. Targeting the c-Rel-Pfkfb3 axis has potential for therapeutic applications in fibrotic disease.
Conflict of interest statement
Figures
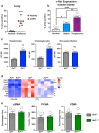

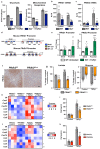
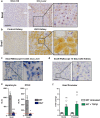
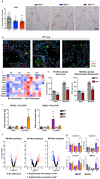







References
Publication types
MeSH terms
Substances
Grants and funding
- MR/R023026/1/MRC_/Medical Research Council/United Kingdom
- A26813/CRUK_/Cancer Research UK/United Kingdom
- MR/K0019494/1/MRC_/Medical Research Council/United Kingdom
- WT107492Z/WT_/Wellcome Trust/United Kingdom
- 26813/CRUK_/Cancer Research UK/United Kingdom
- 20812/VAC_/Versus Arthritis/United Kingdom
- MC_PC_14101/MRC_/Medical Research Council/United Kingdom
- G0401643/MRC_/Medical Research Council/United Kingdom
- L016354/MRC_/Medical Research Council/United Kingdom
- 23390/CRUK_/Cancer Research UK/United Kingdom
- G0700890/MRC_/Medical Research Council/United Kingdom
- 087961/WT_/Wellcome Trust/United Kingdom
- A171196/CRUK_/Cancer Research UK/United Kingdom
- NC/K000748/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- MR/K001949/1/MRC_/Medical Research Council/United Kingdom
- G0900535/MRC_/Medical Research Council/United Kingdom
- A23390/CRUK_/Cancer Research UK/United Kingdom
- MR/L016354/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

