Expression profile and diagnostic value of circRNAs in peripheral blood from patients with systemic lupus erythematosus
- PMID: 33169172
- PMCID: PMC7673322
- DOI: 10.3892/mmr.2020.11639
Expression profile and diagnostic value of circRNAs in peripheral blood from patients with systemic lupus erythematosus
Abstract
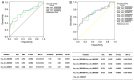
Circular RNAs (circRNAs) have gained attention due to their performance in disease diagnosis. However, the characteristics of circRNAs in peripheral blood from patients with systemic lupus erythematosus (SLE) remain unknown. Therefore, the aim of the present study was to determine the expression profile and diagnostic potential of circRNAs in peripheral blood from patients with SLE. The global circRNA expression in the peripheral blood of patients with SLE and healthy controls (HCs) was detected using a circRNA microarray. Then, the expression levels of three upregulated circRNAs were selected for further validation by reverse transcription‑quantitative PCR (RT‑qPCR) in a training set. Moreover, the diagnostic value of these circRNAs was assessed by constructing a receiver operating characteristic curve, and then verified in a blind testing set. In total, 1,566 circRNAs were identified to be dysregulated between patients with SLE and HCs (≥2 fold change, P<0.05). Furthermore, the RT‑qPCR results were consistent with the microarray data, in that all three selected circRNAs, hsa_circ_0082688, hsa_circ_0082689 and hsa_circ_0008675, were significantly upregulated in patients with SLE (P<0.05). Results from the training set demonstrated that the combination of hsa_circ_0082688‑hsa_circ_0082689 may provide the most beneficial diagnostic potential. Moreover, the blind test results indicated that the combination model of hsa_circ_0082688‑hsa_circ_0082689 could effectively discriminate between patients with SLE from patients with rheumatoid arthritis and HCs, with a sensitivity of 91.30%, a specificity of 78.57% and an accuracy of 82.28%. Moreover, the combination model of hsa_circ_0082688‑hsa_circ_0082689 + anti‑dsDNA could more effectively discriminated the SLE group from the control groups, with a sensitivity of 95.65%, a specificity of 100.00% and an accuracy of 98.73%. In addition, correlation analysis results suggested that all three circRNAs in patients with SLE did not correlate with the SLE disease activity index. In conclusion, the expression levels of hsa_circ_0082688‑hsa_circ_0082689 may serve as potential biomarkers for SLE diagnosis.
Keywords: systemic lupus erythematosus; circular RNAs; microarray assay; peripheral blood.
Figures




Similar articles
-
Identification of circular RNAs hsa_circ_0044235 and hsa_circ_0068367 as novel biomarkers for systemic lupus erythematosus.Int J Mol Med. 2019 Oct;44(4):1462-1472. doi: 10.3892/ijmm.2019.4302. Epub 2019 Aug 5. Int J Mol Med. 2019. PMID: 31432107 Free PMC article.
-
Hsa_circ_0000479 as a Novel Diagnostic Biomarker of Systemic Lupus Erythematosus.Front Immunol. 2019 Sep 24;10:2281. doi: 10.3389/fimmu.2019.02281. eCollection 2019. Front Immunol. 2019. PMID: 31608065 Free PMC article.
-
Microarray expression profile of circular RNAs and mRNAs in children with systemic lupus erythematosus.Clin Rheumatol. 2019 May;38(5):1339-1350. doi: 10.1007/s10067-018-4392-8. Epub 2019 Jan 9. Clin Rheumatol. 2019. PMID: 30628013
-
Significance of circRNAs as biomarkers for systemic lupus erythematosus: a systematic review and meta-analysis.J Int Med Res. 2022 Jul;50(7):3000605221103546. doi: 10.1177/03000605221103546. J Int Med Res. 2022. PMID: 35796516 Free PMC article.
-
The circRNA-miRNA-mRNA regulatory network in plasma and peripheral blood mononuclear cells and the potential associations with the pathogenesis of systemic lupus erythematosus.Clin Rheumatol. 2023 Jul;42(7):1885-1896. doi: 10.1007/s10067-023-06560-5. Epub 2023 Mar 2. Clin Rheumatol. 2023. PMID: 36862342 Review.
Cited by
-
Kaempferol modulates Wnt/ β-catenin pathway to alleviate preeclampsia- induced changes and protect renal and ovarian histomorphology.J Mol Histol. 2024 Dec 7;56(1):36. doi: 10.1007/s10735-024-10321-2. J Mol Histol. 2024. PMID: 39644402
-
circRNA/TLR interaction: key players in immune regulation and autoimmune diseases.Naunyn Schmiedebergs Arch Pharmacol. 2025 May 6. doi: 10.1007/s00210-025-04221-9. Online ahead of print. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 40328911 Review.
-
Potential role of microRNAs in personalized medicine against hepatitis: a futuristic approach.Arch Virol. 2024 Jan 21;169(2):33. doi: 10.1007/s00705-023-05955-8. Arch Virol. 2024. PMID: 38245876 Review.
-
Engineering Large Airways.Adv Exp Med Biol. 2023;1413:121-135. doi: 10.1007/978-3-031-26625-6_7. Adv Exp Med Biol. 2023. PMID: 37195529
-
Chidamide and venetoclax synergistically regulate the Wnt/β-catenin pathway by MYCN/DKK3 in B-ALL.Ann Hematol. 2025 Jan;104(1):489-501. doi: 10.1007/s00277-024-06110-2. Epub 2024 Nov 28. Ann Hematol. 2025. PMID: 39607486 Free PMC article.
References
-
- O'Gorman WE, Hsieh EW, Savig ES, Gherardini PF, Hernandez JD, Hansmann L, Balboni IM, Utz PJ, Bendall SC, Fantl WJ, et al. Single-cell systems-level analysis of human Toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol. 2015;136:1326–1336. doi: 10.1016/j.jaci.2015.04.008. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

