Human Umbilical Cord-Derived Mesenchymal Stem Cell Therapy Effectively Protected the Brain Architecture and Neurological Function in Rat After Acute Traumatic Brain Injury
- PMID: 33169616
- PMCID: PMC7784577
- DOI: 10.1177/0963689720929313
Human Umbilical Cord-Derived Mesenchymal Stem Cell Therapy Effectively Protected the Brain Architecture and Neurological Function in Rat After Acute Traumatic Brain Injury
Abstract
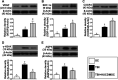
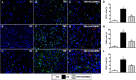
Intracranial hemorrhage from stroke and head trauma elicits a cascade of inflammatory and immune reactions detrimental to neurological integrity and function at cellular and molecular levels. This study tested the hypothesis that human umbilical cord-derived mesenchymal stem cell (HUCDMSC) therapy effectively protected the brain integrity and neurological function in rat after acute traumatic brain injury (TBI). Adult male Sprague-Dawley rats (n = 30) were equally divided into group 1 (sham-operated control), group 2 (TBI), and group 3 [TBI + HUCDMSC (1.2 × 106 cells/intravenous injection at 3 h after TBI)] and euthanized by day 28 after TBI procedure. The results of corner test and inclined plane test showed the neurological function was significantly progressively improved from days 3, 7, 14, and 28 in groups 1 and 3 than in group 2, and group 1 than in group 3 (all P < 0.001). By day 28, brain magnetic resonance imaging brain ischemic volume was significantly increased in group 2 than in group 3 (P < 0.001). The protein expressions of apoptosis [mitochondrial-bax positive cells (Bax)/cleaved-caspase3/cleaved-poly(adenosine diphosphate (ADP)-ribose) polymerase], fibrosis (Smad3 positive cells (Smad3)/transforming growth factor-β), oxidative stress (NADPH Oxidase 1 (NOX-1)/NADPH Oxidase 2 (NOX-2)/oxidized-protein/cytochrome b-245 alpha chain (p22phox)), and brain-edema/deoxyribonucleic acid (DNA)-damaged biomarkers (Aquaporin-4/gamma H2A histone family member X ( (γ-H2AX)) displayed an identical pattern to neurological function among the three groups (all P < 0.0001), whereas the protein expressions of angiogenesis biomarkers (vascular endothelial growth factor/stromal cell-derived factor-1α/C-X-C chemokine receptor type 4 (CXCR4)) significantly increased from groups 1 to 3 (all P < 0.0001). The cellular expressions of inflammatory biomarkers (cluster of differentiation 14 (+) cells (CD14+)/glial fibrillary acidic protein positive cells (GFAP+)/ a member of a new family of EGF-TM7 molecules positive cells (F4/80+)) and DNA-damaged parameter (γ-H2AX) exhibited an identical pattern, whereas cellular expressions of neural integrity (hexaribonucleotide Binding Protein-3 positive cells (NeuN+)/nestin+/doublecortin+) exhibited an opposite pattern of neurological function among the three groups (all P < 0.0001). Xenogeneic HUCDMSC therapy was safe and it significantly preserved neurological function and brain architecture in rat after TBI.
Keywords: apoptosis; inflammation; neurological function; oxidative stress; traumatic brain injury; xenogeneic cell therapy.
Conflict of interest statement
Figures






Similar articles
-
Dose-dependent benefits of iron-magnetic nanoparticle-coated human umbilical-derived mesenchymal stem cell treatment in rat intracranial hemorrhage model.Stem Cell Res Ther. 2022 Jun 21;13(1):265. doi: 10.1186/s13287-022-02939-4. Stem Cell Res Ther. 2022. PMID: 35729660 Free PMC article.
-
Umbilical cord-derived MSC and hyperbaric oxygen therapy effectively protected the brain in rat after acute intracerebral haemorrhage.J Cell Mol Med. 2021 Jun;25(12):5640-5654. doi: 10.1111/jcmm.16577. Epub 2021 May 2. J Cell Mol Med. 2021. PMID: 33938133 Free PMC article.
-
Intravenous administration of xenogenic adipose-derived mesenchymal stem cells (ADMSC) and ADMSC-derived exosomes markedly reduced brain infarct volume and preserved neurological function in rat after acute ischemic stroke.Oncotarget. 2016 Nov 15;7(46):74537-74556. doi: 10.18632/oncotarget.12902. Oncotarget. 2016. PMID: 27793019 Free PMC article.
-
Treating childhood traumatic brain injury with autologous stem cell therapy.Expert Opin Biol Ther. 2018 May;18(5):515-524. doi: 10.1080/14712598.2018.1439473. Epub 2018 Feb 15. Expert Opin Biol Ther. 2018. PMID: 29421958 Free PMC article. Review.
-
Potential of mesenchymal stem cells alone, or in combination, to treat traumatic brain injury.CNS Neurosci Ther. 2020 Jun;26(6):616-627. doi: 10.1111/cns.13300. Epub 2020 Mar 10. CNS Neurosci Ther. 2020. PMID: 32157822 Free PMC article. Review.
Cited by
-
Ischemic Stroke: Pathophysiology and Evolving Treatment Approaches.Neurosci Insights. 2024 Oct 22;19:26331055241292600. doi: 10.1177/26331055241292600. eCollection 2024. Neurosci Insights. 2024. PMID: 39444789 Free PMC article. Review.
-
Molecular Pathogenesis of Ischemic and Hemorrhagic Strokes: Background and Therapeutic Approaches.Int J Mol Sci. 2024 Jun 7;25(12):6297. doi: 10.3390/ijms25126297. Int J Mol Sci. 2024. PMID: 38928006 Free PMC article. Review.
-
Stroke: Molecular mechanisms and therapies: Update on recent developments.Neurochem Int. 2023 Jan;162:105458. doi: 10.1016/j.neuint.2022.105458. Epub 2022 Nov 30. Neurochem Int. 2023. PMID: 36460240 Free PMC article. Review.
-
Exosomes Derived From Mesenchymal Stem Cells: Novel Effects in the Treatment of Ischemic Stroke.Front Neurosci. 2022 May 2;16:899887. doi: 10.3389/fnins.2022.899887. eCollection 2022. Front Neurosci. 2022. PMID: 35585925 Free PMC article. Review.
-
Dose-dependent benefits of iron-magnetic nanoparticle-coated human umbilical-derived mesenchymal stem cell treatment in rat intracranial hemorrhage model.Stem Cell Res Ther. 2022 Jun 21;13(1):265. doi: 10.1186/s13287-022-02939-4. Stem Cell Res Ther. 2022. PMID: 35729660 Free PMC article.
References
-
- Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008;371(9624):1612–1623. - PubMed
-
- Hu HH, Sheng WY, Chu FL, Lan CF, Chiang BN. Incidence of stroke in Taiwan. Stroke. 1992;23(9):1237–1241. - PubMed
-
- Hung TP. [Clinical aspects of cerebral hemorrhage]. Taiwan Yi Xue Hui Za Zhi. 1988;87(3):261–723. - PubMed
-
- Carlo DA. Human and economic burden of stroke. Age Ageing. 2009;38(1):4–5. - PubMed
-
- Masdeu JC, Rubino FA. Management of lobar intracerebral hemorrhage: medical or surgical. Neurol. 1984;34(3):381–383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

