Etiology of Severe Community-Acquired Pneumonia in Adults Based on Metagenomic Next-Generation Sequencing: A Prospective Multicenter Study
- PMID: 33170499
- PMCID: PMC7652912
- DOI: 10.1007/s40121-020-00353-y
Etiology of Severe Community-Acquired Pneumonia in Adults Based on Metagenomic Next-Generation Sequencing: A Prospective Multicenter Study
Abstract
Background: Metagenomic next-generation sequencing (mNGS) of bronchoalveolar lavage fluid (BALF) has the potential to improve the pathogen identification in severe community-acquired pneumonia (SCAP).
Methods: In this 1.5-year, multicenter, prospective study, we investigated the usefulness of mNGS of BALF for identifying pathogens of SCAP in hospitalized adults, comparing it with other laboratory methods.
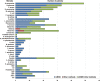
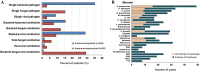
Results: Of 329 SCAP adults, a microbial etiology was established in 304 cases (92.4%). The overall microbial yield was 90.3% for mNGS versus 39.5% for other methods (P < 0.05). The most frequently detected pathogens in immunocompetent patients were Streptococcus pneumoniae (14.8%), rhinovirus (9.8%), Haemophilus influenzae (9.1%), Staphylococcus aureus (8.7%), and Chlamydia psittaci (8.0%), while in immunocompromised patients they were Pneumocystis jirovecii (44.6%), Klebsiella pneumoniae (18.5%), Streptococcus pneumoniae (15.4%), Haemophilus influenzae (13.8%), and Pseudomonas aeruginosa (13.8%). Notably, novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified from two patients solely by mNGS in January 2020; uncommon pathogens including Orientia tsutsugamushi and Nocardia otitidiscaviarum were identified from one patient, respectively. Furthermore, mixed infections were detected in 56.8% of the patients.
Conclusions: A high microbial detection rate was achieved in SCAP adults using mNGS testing of BALF. The most frequently detected pathogens of SCAP differed between immunocompetent and immunocompromised patients. mNGS testing may be an powerful tool for early identification of potential pathogens for SCAP to initiate a precise antimicrobial therapy.
Keywords: Etiology; Metagenomic next-generation sequencing; Severe community-acquired pneumonia.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

