Deubiquitinase USP7 contributes to the pathogenicity of spinal and bulbar muscular atrophy
- PMID: 33170804
- PMCID: PMC7773404
- DOI: 10.1172/JCI134565
Deubiquitinase USP7 contributes to the pathogenicity of spinal and bulbar muscular atrophy
Abstract
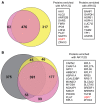
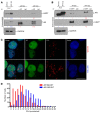
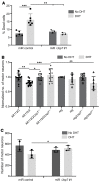
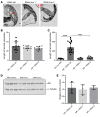
Polyglutamine (polyQ) diseases are devastating, slowly progressing neurodegenerative conditions caused by expansion of polyQ-encoding CAG repeats within the coding regions of distinct, unrelated genes. In spinal and bulbar muscular atrophy (SBMA), polyQ expansion within the androgen receptor (AR) causes progressive neuromuscular toxicity, the molecular basis of which is unclear. Using quantitative proteomics, we identified changes in the AR interactome caused by polyQ expansion. We found that the deubiquitinase USP7 preferentially interacts with polyQ-expanded AR and that lowering USP7 levels reduced mutant AR aggregation and cytotoxicity in cell models of SBMA. Moreover, USP7 knockdown suppressed disease phenotypes in SBMA and spinocerebellar ataxia type 3 (SCA3) fly models, and monoallelic knockout of Usp7 ameliorated several motor deficiencies in transgenic SBMA mice. USP7 overexpression resulted in reduced AR ubiquitination, indicating the direct action of USP7 on AR. Using quantitative proteomics, we identified the ubiquitinated lysine residues on mutant AR that are regulated by USP7. Finally, we found that USP7 also differentially interacts with mutant Huntingtin (HTT) protein in striatum and frontal cortex of a knockin mouse model of Huntington's disease. Taken together, our findings reveal a critical role for USP7 in the pathophysiology of SBMA and suggest a similar role in SCA3 and Huntington's disease.
Keywords: Neurodegeneration; Neuromuscular disease; Neuroscience; Ubiquitin-proteosome system.
Conflict of interest statement
Figures





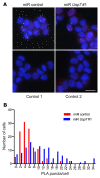
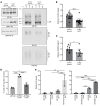
References
-
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

