Quick COVID-19 Healers Sustain Anti-SARS-CoV-2 Antibody Production
- PMID: 33171099
- PMCID: PMC7608032
- DOI: 10.1016/j.cell.2020.10.051
Quick COVID-19 Healers Sustain Anti-SARS-CoV-2 Antibody Production
Abstract
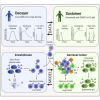
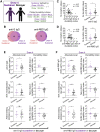
Antibodies are key immune effectors that confer protection against pathogenic threats. The nature and longevity of the antibody response to SARS-CoV-2 infection are not well defined. We charted longitudinal antibody responses to SARS-CoV-2 in 92 subjects after symptomatic COVID-19. Antibody responses to SARS-CoV-2 are unimodally distributed over a broad range, with symptom severity correlating directly with virus-specific antibody magnitude. Seventy-six subjects followed longitudinally to ∼100 days demonstrated marked heterogeneity in antibody duration dynamics. Virus-specific IgG decayed substantially in most individuals, whereas a distinct subset had stable or increasing antibody levels in the same time frame despite similar initial antibody magnitudes. These individuals with increasing responses recovered rapidly from symptomatic COVID-19 disease, harbored increased somatic mutations in virus-specific memory B cell antibody genes, and had persistent higher frequencies of previously activated CD4+ T cells. These findings illuminate an efficient immune phenotype that connects symptom clearance speed to differential antibody durability dynamics.
Keywords: COVID-19; SARS-CoV-2; SHM; durability; germinal center; serology; severity; somatic hypermutation; symptom duration.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures













References
-
- Amanna I.J., Carlson N.E., Slifka M.K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 2007;357:1903–1915. - PubMed
-
- Balaz M., Becker A.S., Balazova L., Straub L., Muller J., Gashi G., Maushart C.I., Sun W., Dong H., Moser C. Inhibition of Mevalonate Pathway Prevents Adipocyte Browning in Mice and Men by Affecting Protein Prenylation. Cell Metab. 2019;29:901–916.e8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19 AI135995/AI/NIAID NIH HHS/United States
- R01 AI139538/AI/NIAID NIH HHS/United States
- U19 AI142790/AI/NIAID NIH HHS/United States
- R01 AI146779/AI/NIAID NIH HHS/United States
- T32 GM008313/GM/NIGMS NIH HHS/United States
- R21 AI137940/AI/NIAID NIH HHS/United States
- R01 AI170715/AI/NIAID NIH HHS/United States
- T32 AI007245/AI/NIAID NIH HHS/United States
- T32 AI007306/AI/NIAID NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32 AI007512/AI/NIAID NIH HHS/United States
- P01 AI165072/AI/NIAID NIH HHS/United States
- DP2 DA040254/DA/NIDA NIH HHS/United States
- R01 AI121394/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

