Changes in Phenolics and Fatty Acids Composition and Related Gene Expression during the Development from Seed to Leaves of Three Cultivated Cardoon Genotypes
- PMID: 33171628
- PMCID: PMC7695130
- DOI: 10.3390/antiox9111096
Changes in Phenolics and Fatty Acids Composition and Related Gene Expression during the Development from Seed to Leaves of Three Cultivated Cardoon Genotypes
Abstract
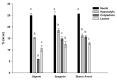
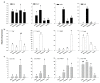
Cultivated cardoon (Cynara cardunculus var. altilis) has long been used as a food and medicine remedy and nowadays is considered a functional food. Its leaf bioactive compounds are mostly represented by chlorogenic acids and coumaroyl derivatives, known for their nutritional value and bioactivity. Having antioxidant and hepatoprotective properties, these molecules are used for medicinal purposes. Apart from the phenolic compounds in green tissues, cultivated cardoon is also used for the seed oil, having a composition suitable for the human diet, but also valuable as feedstock for the production of biofuel and biodegradable bioplastics. Given the wide spectrum of valuable cardoon molecules and their numerous industrial applications, a detailed characterization of different organs and tissues for their metabolic profiles as well as an extensive transcriptional analysis of associated key biosynthetic genes were performed to provide a deeper insight into metabolites biosynthesis and accumulation sites. This study aimed to provide a comprehensive analysis of the phenylpropanoids profile through UHPLC-Q-Orbitrap HRMS analysis, of fatty acids content through GC-MS analysis, along with quantitative transcriptional analyses by qRT-PCR of hydroxycinnamoyl-quinate transferase (HQT), stearic acid desaturase (SAD), and fatty acid desaturase (FAD) genes in seeds, hypocotyls, cotyledons and leaves of the cardoon genotypes "Spagnolo", "Bianco Avorio", and "Gigante". Both oil yield and total phenols accumulation in all the tissues and organs indicated higher production in "Bianco Avorio" and "Spagnolo" than in "Gigante". Antioxidant activity evaluation by DPPH, ABTS, and FRAP assays mirrored total phenols content. Overall, this study provides a detailed analysis of tissue composition of cardoon, enabling to elucidate value-added product accumulation and distribution during plant development and hence contributing to better address and optimize the sustainable use of this natural resource. Besides, our metabolic and transcriptional screening could be useful to guide the selection of superior genotypes.
Keywords: antioxidant; cardoon; chlorogenic acid; fatty acids; multipurpose plant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bianco V.V. Present situation and future potential of artichoke in the mediterranean basin. Acta Hortic. 2005;681:39–58. doi: 10.17660/ActaHortic.2005.681.2. - DOI
-
- Ciancolini A., Alignan M., Pagnotta M.A., Vilarem G., Crinò P. Selection of Italian cardoon genotypes as industrial crop for biomass and polyphenol production. Ind. Crop. Prod. 2013;51:145–151. doi: 10.1016/j.indcrop.2013.08.069. - DOI
-
- Foti S., Mauromicale G., Raccuia S.A., Fallico B., Fanella F. Possible alternative utilization of Cynara spp. I. Biomass, grain yield and chemical composition of grain. Ind. Crops Prod. 1999;10:219–228. doi: 10.1016/S0926-6690(99)00026-6. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

