Bile Acid Signal Molecules Associate Temporally with Respiratory Inflammation and Microbiome Signatures in Clinically Stable Cystic Fibrosis Patients
- PMID: 33172004
- PMCID: PMC7694639
- DOI: 10.3390/microorganisms8111741
Bile Acid Signal Molecules Associate Temporally with Respiratory Inflammation and Microbiome Signatures in Clinically Stable Cystic Fibrosis Patients
Abstract
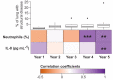
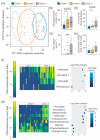
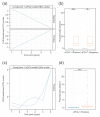
Cystic fibrosis (CF) is a congenital disorder resulting in a multisystemic impairment in ion homeostasis. The subsequent alteration of electrochemical gradients severely compromises the function of the airway epithelia. These functional changes are accompanied by recurrent cycles of inflammation-infection that progressively lead to pulmonary insufficiency. Recent developments have pointed to the existence of a gut-lung axis connection, which may modulate the progression of lung disease. Molecular signals governing the interplay between these two organs are therefore candidate molecules requiring further clinical evaluation as potential biomarkers. We demonstrate a temporal association between bile acid (BA) metabolites and inflammatory markers in bronchoalveolar lavage fluid (BALF) from clinically stable children with CF. By modelling the BALF-associated microbial communities, we demonstrate that profiles enriched in operational taxonomic units assigned to supraglottic taxa and opportunistic pathogens are closely associated with inflammatory biomarkers. Applying regression analyses, we also confirmed a linear link between BA concentration and pathogen abundance in BALF. Analysis of the time series data suggests that the continuous detection of BAs in BALF is linked to differential ecological succession trajectories of the lung microbiota. Our data provide further evidence supporting a role for BAs in the early pathogenesis and progression of CF lung disease.
Keywords: bile acids; cystic fibrosis; gut–lung axis; inflammation; lung microbiota.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Ranganathan S.C., Hall G.L., Sly P.D., Stick S.M., Douglas T.A., Australian Respiratory Early Surveillance Team for Cystic, F Early Lung Disease in Infants and Preschool Children with Cystic Fibrosis. What Have We Learned and What Should We Do about It? Am. J. Respir. Crit. Care Med. 2017;195:1567–1575. doi: 10.1164/rccm.201606-1107CI. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

