New Insights on Molecular Mechanism of Hepatitis B Virus Covalently Closed Circular DNA Formation
- PMID: 33172220
- PMCID: PMC7694973
- DOI: 10.3390/cells9112430
New Insights on Molecular Mechanism of Hepatitis B Virus Covalently Closed Circular DNA Formation
Abstract
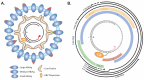
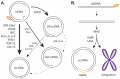
The chronic factor of the Hepatitis B Virus (HBV), specifically the covalently closed circular DNA (cccDNA), is a highly stable and active viral episomal genome established in the livers of chronic hepatitis B patients as a constant source of disease. Being able to target and eliminate cccDNA is the end goal for a genuine cure for HBV. Yet how HBV cccDNA is formed from the viral genomic relaxed circular DNA (rcDNA) and by what host factors had been long-standing research questions. It is generally acknowledged that HBV hijacks cellular functions to turn the open circular DNA conformation of rcDNA into cccDNA through DNA repair mechanisms. With great efforts from the HBV research community, there have been several recent leaps in our understanding of cccDNA formation. It is our goal in this review to analyze the recent reports showing evidence of cellular factor's involvement in the molecular pathway of cccDNA biosynthesis.
Keywords: DNA repair; covalently closed circular DNA; hepatitis B virus; replication.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

