Proteogenomic Analysis of Salivary Adenoid Cystic Carcinomas Defines Molecular Subtypes and Identifies Therapeutic Targets
- PMID: 33172898
- PMCID: PMC7854509
- DOI: 10.1158/1078-0432.CCR-20-1192
Proteogenomic Analysis of Salivary Adenoid Cystic Carcinomas Defines Molecular Subtypes and Identifies Therapeutic Targets
Erratum in
-
Correction: Proteogenomic Analysis of Salivary Adenoid Cystic Carcinomas Defines Molecular Subtypes and Identifies Therapeutic Targets.Clin Cancer Res. 2023 Jul 14;29(14):2737. doi: 10.1158/1078-0432.CCR-23-1697. Clin Cancer Res. 2023. PMID: 37449360 No abstract available.
Abstract
Purpose: Salivary gland adenoid cystic carcinoma (ACC) has heterogeneous clinical behavior. Currently, all patients are treated uniformly, and no standard-of-care systemic therapy exists for metastatic ACC. We conducted an integrated proteogenomic analyses of ACC tumors to identify dysregulated pathways and propose a classification with therapeutic implications.
Experimental design: RNA/DNA sequencing of 54 flash-frozen salivary ACCs and reverse phase protein array (RPPA) in 38 specimens were performed, with validation by Western blotting and/or IHC. Three independent ACC cohorts were used for validation.
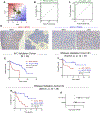
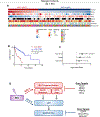
Results: Both unbiased RNA sequencing (RNA-seq) and RPPA analysis revealed two molecular subtypes: ACC-I (37%) and ACC-II (63%). ACC-I had strong upregulation of MYC, MYC target genes, and mRNA splicing, enrichment of NOTCH-activating mutations, and dramatically worse prognosis. ACC-II exhibited upregulation of TP63 and receptor tyrosine kinases (AXL, MET, and EGFR) and less aggressive clinical course. TP63 and MYC were sufficient to assign tumors to ACC subtypes, which was validated in one independent cohort by IHC and two additional independent cohorts by RNA-seq. Furthermore, IHC staining for MYC and P63 protein levels can be used to identify ACC subtypes, enabling rapid clinical deployment to guide therapeutic decisions. Our data suggest a model in which ACC-I is driven by MYC signaling through either NOTCH mutations or direct amplification, which in turn suppress P63 signaling observed in ACC-II, producing unique therapeutic vulnerabilities for each subtype.
Conclusions: Cooccurrence of multiple actionable protein/pathways alterations in each subtype indicates unique therapeutic vulnerabilities and opportunities for optimal combination therapy for this understudied and heterogeneous disease.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures
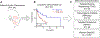



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

