Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans
- PMID: 33172935
- PMCID: PMC7857398
- DOI: 10.1126/science.abe5901
Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans
Abstract
Animal experiments have shown that nonhuman primates, cats, ferrets, hamsters, rabbits, and bats can be infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In addition, SARS-CoV-2 RNA has been detected in felids, mink, and dogs in the field. Here, we describe an in-depth investigation using whole-genome sequencing of outbreaks on 16 mink farms and the humans living or working on these farms. We conclude that the virus was initially introduced by humans and has since evolved, most likely reflecting widespread circulation among mink in the beginning of the infection period, several weeks before detection. Despite enhanced biosecurity, early warning surveillance, and immediate culling of animals in affected farms, transmission occurred between mink farms in three large transmission clusters with unknown modes of transmission. Of the tested mink farm residents, employees, and/or individuals with whom they had been in contact, 68% had evidence of SARS-CoV-2 infection. Individuals for which whole genomes were available were shown to have been infected with strains with an animal sequence signature, providing evidence of animal-to-human transmission of SARS-CoV-2 within mink farms.
Copyright © 2021, American Association for the Advancement of Science.
Figures
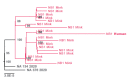


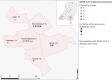
References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
-
- Oude Munnink B. B., Nieuwenhuijse D. F., Stein M., O’Toole Á., Haverkate M., Mollers M., Kamga S. K., Schapendonk C., Pronk M., Lexmond P., van der Linden A., Bestebroer T., Chestakova I., Overmars R. J., van Nieuwkoop S., Molenkamp R., van der Eijk A. A., GeurtsvanKessel C., Vennema H., Meijer A., Rambaut A., van Dissel J., Sikkema R. S., Timen A., Koopmans M.; Dutch-Covid-19 response team , Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat. Med. 26, 1405–1410 (2020). 10.1038/s41591-020-0997-y - DOI - PubMed
-
- Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., Hu Y., Tao Z. W., Tian J. H., Pei Y. Y., Yuan M. L., Zhang Y. L., Dai F. H., Liu Y., Wang Q. M., Zheng J. J., Xu L., Holmes E. C., Zhang Y. Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
-
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E. C., Hughes A. C., Bi Y., Shi W., A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 30, 3896 (2020). 10.1016/j.cub.2020.09.030 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

