Gut microbiota and metabolic health among overweight and obese individuals
- PMID: 33173145
- PMCID: PMC7655835
- DOI: 10.1038/s41598-020-76474-8
Gut microbiota and metabolic health among overweight and obese individuals
Abstract
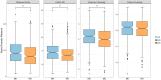
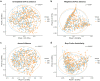
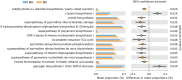
Although obesity is associated with numerous diseases, the risks of disease may depend on metabolic health. Associations between the gut microbiota, obesity, and metabolic syndrome have been reported, but differences in microbiomes according to metabolic health in the obese population have not been explored in previous studies. Here, we investigated the composition of gut microbiota according to metabolic health status in obese and overweight subjects. A total of 747 overweight or obese adults were categorized by metabolic health status, and their fecal microbiota were profiled using 16S ribosomal RNA gene sequencing. We classified these adults into a metabolically healthy group (MH, N = 317) without any components of metabolic syndrome or a metabolically unhealthy group (MU, N = 430) defined as having at least one metabolic abnormality. The phylogenetic and non-phylogenetic alpha diversity for gut microbiota were lower in the MU group than the MH group, and there were significant differences in gut microbiota bacterial composition between the two groups. We found that the genus Oscillospira and the family Coriobacteriaceae were associated with good metabolic health in the overweight and obese populations. This is the first report to describe gut microbial diversity and composition in metabolically healthy and unhealthy overweight and obese individuals. Modulation of the gut microbiome may help prevent metabolic abnormalities in the obese population.
Conflict of interest statement
The authors declare no competing interests.
Figures
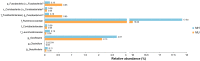

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

