Insulin-promoted mobilization of GLUT4 from a perinuclear storage site requires RAB10
- PMID: 33175605
- PMCID: PMC8098823
- DOI: 10.1091/mbc.E20-06-0356
Insulin-promoted mobilization of GLUT4 from a perinuclear storage site requires RAB10
Abstract
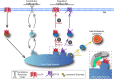
Insulin controls glucose uptake into muscle and fat cells by inducing a net redistribution of glucose transporter 4 (GLUT4) from intracellular storage to the plasma membrane (PM). The TBC1D4-RAB10 signaling module is required for insulin-stimulated GLUT4 translocation to the PM, although where it intersects GLUT4 traffic was unknown. Here we demonstrate that TBC1D4-RAB10 functions to control GLUT4 mobilization from a trans-Golgi network (TGN) storage compartment, establishing that insulin, in addition to regulating the PM proximal effects of GLUT4-containing vesicles docking to and fusion with the PM, also directly regulates the behavior of GLUT4 deeper within the cell. We also show that GLUT4 is retained in an element/domain of the TGN from which newly synthesized lysosomal proteins are targeted to the late endosomes and the ATP7A copper transporter is translocated to the PM by elevated copper. Insulin does not mobilize ATP7A nor does copper mobilize GLUT4, and RAB10 is not required for copper-elicited ATP7A mobilization. Consequently, GLUT4 intracellular sequestration and mobilization by insulin is achieved, in part, through utilizing a region of the TGN devoted to specialized cargo transport in general rather than being specific for GLUT4. Our results define the GLUT4-containing region of the TGN as a sorting and storage site from which different cargo are mobilized by distinct signals through unique molecular machinery.
Figures
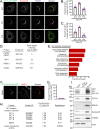

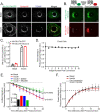
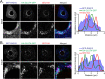
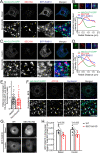

References
-
- Bai L, Wang Y, Fan J, Chen Y, Ji W, Qu A, Xu P, James DE, Xu T (2007). Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metab 5, 47–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

