P.F508del editing in cells from cystic fibrosis patients
- PMID: 33175893
- PMCID: PMC7657551
- DOI: 10.1371/journal.pone.0242094
P.F508del editing in cells from cystic fibrosis patients
Abstract
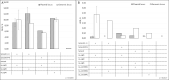
Development of genome editing methods created new opportunities for the development of etiology-based therapies of hereditary diseases. Here, we demonstrate that CRISPR/Cas9 can correct p.F508del mutation in the CFTR gene in the CFTE29o- cells and induced pluripotent stem cells (iPSCs) derived from patients with cystic fibrosis (CF). We used several combinations of Cas9, sgRNA and ssODN and measured editing efficiency in the endogenous CFTR gene and in the co-transfected plasmid containing the CFTR locus with the p.F508del mutation. The non-homologous end joining (NHEJ) frequency in the CFTR gene in the CFTE29o- cells varied from 1.25% to 2.54% of alleles. The best homology-directed repair (HDR) frequency in the endogenous CFTR locus was 1.42% of alleles. In iPSCs, the NHEJ frequency in the CFTR gene varied from 5.5% to 12.13% of alleles. The best HDR efficacy was 2.38% of alleles. Our results show that p.F508del mutation editing using CRISPR/Cas9 in CF patient-derived iPSCs is a relatively rare event and subsequent cell selection and cultivation should be carried out.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Cystic Fibrosis Foundation. Patient registry: annual data report, 2017. Bethesda, Maryland. 2018. https://www.cff.org/Research/Researcher-Resources/Patient-Registry/2017-.... Accessed 20 May 2020.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

