Bipotent Progenitors Do Not Require Androgen Receptor for Luminal Specification during Prostate Organogenesis
- PMID: 33176121
- PMCID: PMC7664048
- DOI: 10.1016/j.stemcr.2020.10.004
Bipotent Progenitors Do Not Require Androgen Receptor for Luminal Specification during Prostate Organogenesis
Abstract
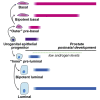
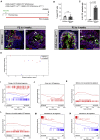
Androgen receptor (AR) plays a fundamental role in most aspects of adult prostate homeostasis, and anti-androgen therapy represents the cornerstone of prostate cancer treatment. However, early prostate organogenesis takes place during pre-pubertal stages when androgen levels are low, raising the possibility that AR function is more limited during prostate development. Here, we use inducible AR deletion and lineage tracing in genetically engineered mice to show that basal and luminal epithelial progenitors do not require cell-autonomous AR activity during prostate development. We also demonstrate the existence of a transient bipotent luminal progenitor that can generate luminal and basal progeny, yet is also independent of AR function. Furthermore, molecular analyses of AR-deleted luminal cells isolated from developing prostates indicate their similarity to wild-type cells. Our findings suggest that low androgen levels correlate with luminal plasticity in prostate development and may have implications for understanding how AR inhibition promotes lineage plasticity in prostate cancer.
Keywords: androgen receptor; lineage tracing; luminal progenitor; plasticity; prostate organogenesis.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

