Avian Immunome DB: an example of a user-friendly interface for extracting genetic information
- PMID: 33176685
- PMCID: PMC7661159
- DOI: 10.1186/s12859-020-03764-3
Avian Immunome DB: an example of a user-friendly interface for extracting genetic information
Erratum in
-
Correction to: Avian Immunome DB: an example of a user‑friendly interface for extracting genetic information.BMC Bioinformatics. 2021 Sep 30;22(1):472. doi: 10.1186/s12859-021-04388-x. BMC Bioinformatics. 2021. PMID: 34592921 Free PMC article. No abstract available.
Abstract
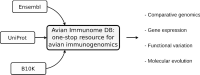
Background: Genomic and genetic studies often require a target list of genes before conducting any hypothesis testing or experimental verification. With the ever-growing number of sequenced genomes and a variety of different annotation strategies, comes the potential for ambiguous gene symbols, making it cumbersome to capture the "correct" set of genes. In this article, we present and describe the Avian Immunome DB (AVIMM) for easy gene property extraction as exemplified by avian immune genes. The avian immune system is characterised by a cascade of complex biological processes underlaid by more than 1000 different genes. It is a vital trait to study particularly in birds considering that they are a significant driver in spreading zoonotic diseases. With the completion of phase II of the B10K ("Bird 10,000 Genomes") consortium's whole-genome sequencing effort, we have included 363 annotated bird genomes in addition to other publicly available bird genome data which serve as a valuable foundation for AVIMM.
Construction and content: A relational database with avian immune gene evidence from Gene Ontology, Ensembl, UniProt and the B10K consortium has been designed and set up. The foundation stone or the "seed" for the initial set of avian immune genes is based on the well-studied model organism chicken (Gallus gallus). Gene annotations, different transcript isoforms, nucleotide sequences and protein information, including amino acid sequences, are included. Ambiguous gene names (symbols) are resolved within the database and linked to their canonical gene symbol. AVIMM is supplemented by a command-line interface and a web front-end to query the database.
Utility and discussion: The internal mapping of unique gene symbol identifiers to canonical gene symbols allows for an ambiguous gene property search. The database is organised within core and feature tables, which makes it straightforward to extend for future purposes. The database design is ready to be applied to other taxa or biological processes. Currently, the database contains 1170 distinct avian immune genes with canonical gene symbols and 612 synonyms across 363 bird species. While the command-line interface readily integrates into bioinformatics pipelines, the intuitive web front-end with download functionality offers sophisticated search functionalities and tracks the origin for each record. AVIMM is publicly accessible at https://avimm.ab.mpg.de .
Keywords: Avian; B10K; Genomics; Immunogenomics; Immunology; Immunome; Trait database.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- GenBank and WGS Statistics. https://www.ncbi.nlm.nih.gov/genbank/statistics/. Accessed 13 May 2020
-
- Selzer PM, Marhöfer RJ, Koch O. Biological databases. In: Selzer PM, Marhöfer RJ, Koch O, editors. Applied bioinformatics: an Introduction. Cham: Springer; 2018. pp. 13–34.
-
- Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, Davidson C, Dodiya KJ, Gall A, Girón CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Laird MR, Lavidas I, Liu Z, Loveland JE, Marugán JC, Maurel T, McMahon AC, Moore B, Morales J, Mudge JM, Nuhn M, Ogeh D, Parker A, Parton A, Patricio M, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sparrow H, Stapleton E, Szuba M, Taylor K, Threadgold G, Thormann A, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Yates AD, Zerbino DR, Flicek P. Ensembl. Nucleic Acids Res. 2019;47(D1):745–51. doi: 10.1093/nar/gky1113. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

