ATF4 activation promotes hepatic mitochondrial dysfunction by repressing NRF1-TFAM signalling in alcoholic steatohepatitis
- PMID: 33177163
- PMCID: PMC8110597
- DOI: 10.1136/gutjnl-2020-321548
ATF4 activation promotes hepatic mitochondrial dysfunction by repressing NRF1-TFAM signalling in alcoholic steatohepatitis
Abstract
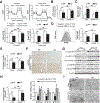
Objective: Mitochondrial dysfunction plays a dominant role in the pathogenesis of alcoholic liver disease (ALD); however, the underlying mechanisms remain to be fully understood. We previously found that hepatic activating transcription factor 4 (ATF4) activation was associated with mitochondrial dysfunction in ALD. This study aimed to investigate the function and mechanism of ATF4 in alcohol-induced hepatic mitochondrial dysfunction.
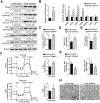
Design: ATF4 activation was detected in the livers of patients with severe alcoholic hepatitis (AH). The role of ATF4 and mitochondrial transcription factor A (TFAM) in alcohol-induced liver damage was determined in hepatocyte-specific ATF4 knockout mice and liver-specific TFAM overexpression mice, respectively.
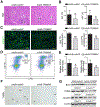
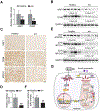
Results: Hepatic PERK-eIF2α-ATF4 ER stress signalling was upregulated in patients with AH. Hepatocyte-specific ablation of ATF4 in mice ameliorated alcohol-induced steatohepatitis. ATF4 ablation also attenuated alcohol-impaired mitochondrial biogenesis and respiratory function along with the restoration of TFAM. Cell studies confirmed that TFAM expression was negatively regulated by ATF4. TFAM silencing in hepatoma cells abrogated the protective effects of ATF4 knockdown on ethanol-mediated mitochondrial dysfunction and cell death. Moreover, hepatocyte-specific TFAM overexpression in mice attenuated alcohol-induced mitochondrial dysfunction and liver damage. Mechanistic studies revealed that ATF4 repressed the transcription activity of nuclear respiratory factor 1 (NRF1), a key regulator of TFAM, through binding to its promoter region. Clinical relevance among ATF4 activation, NRF1-TFAM pathway disruption and mitochondrial dysfunction was validated in the livers of patients with AH.
Conclusion: This study demonstrates that hepatic ATF4 plays a pathological role in alcohol-induced mitochondrial dysfunction and liver injury by disrupting the NRF1-TFAM pathway.
Keywords: alcoholic liver disease; energy metabolism; hepatocyte.
© Author(s) (or their employer(s)) 2021. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
-
- Seitz HK, Bataller R, Cortez-Pinto H, et al.Alcoholic liver disease. Nature reviews Disease primers 2018;4:16. - PubMed
-
- Thursz M, Kamath PS, Mathurin P, et al.Alcohol-related liver disease: Areas of consensus, unmet needs and opportunities for further study. Journal of hepatology 2019;70:521–530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
