cDNA-Derived RNA Phage Assembly Reveals Critical Residues in the Maturation Protein of the Pseudomonas aeruginosa Leviphage PP7
- PMID: 33177196
- PMCID: PMC7925092
- DOI: 10.1128/JVI.01643-20
cDNA-Derived RNA Phage Assembly Reveals Critical Residues in the Maturation Protein of the Pseudomonas aeruginosa Leviphage PP7
Abstract
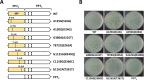
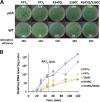
PP7 is a leviphage, with a single-stranded RNA genome, that infects Pseudomonas aeruginosa PAO1. A reverse genetic system for PP7 was previously created by using reverse-transcribed cDNA (PP7O) from a virion-derived RNA genome. Here, we have found that the PP7O cDNA contained 20 nucleotide differences from the PP7 genome sequence deposited in the database. We created another reverse genetic system exploiting chemically synthesized cDNA (PP7S) based on the database sequence. Unlike PP7O, which yielded infectious PP7 virions, PP7S-derived particles were incapable of plaque formation on PAO1 cells, which was restored in the PAO1 cells expressing the maturation protein (MP) from PP7O Using this reverse genetic system, we revealed two amino acid residues involved in the known roles of MP (i.e., adsorption and genome replication), fortuitously providing a lesson that the viral RNA genome sequencing needs functional verification, possibly by a reverse genetic system.IMPORTANCE The biological significance of RNA phages has been largely ignored, ironically, because few studies have focused on RNA phages. As an initial attempt to properly represent RNA phages in the phageome, we previously created, by using reverse-transcribed cDNA, a reverse genetic system for the small RNA phage PP7, which infects the opportunistic human pathogen Pseudomonas aeruginosa We report another system by using chemically synthesized cDNA based on the database genome that has 20 nucleotide differences from the previous cDNA. Investigation of those cDNA-derived phage virions revealed that two amino acids of the maturation protein are crucial for the normal phage lifecycle at different steps. Our study provides insight into the molecular basis for the RNA phage lifecycle and a lesson that the RNA genome sequencing needs to be carefully validated by cDNA-based phage assembly systems.
Keywords: PP7; Pseudomonas aeruginosa; RNA phage; cDNA; infectivity; maturation protein.
Copyright © 2021 American Society for Microbiology.
Figures




References
-
- Palladini A, Thrane S, Janitzek CM, Pihl J, Clemmensen SB, de Jongh WA, Clausen TM, Nicoletti G, Landuzzi L, Penichet ML, Balboni T, Ianzano ML, Giusti V, Theander TG, Nielsen MA, Salanti A, Lollini P-L, Nanni P, Sander AF. 2018. Virus-like particle display of HER2 induces potent anti-cancer responses. Oncoimmunology 7:e1408749. doi:10.1080/2162402X.2017.1408749. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

